Latent path analysis
In this tutorial, we are going to use lavaan for latent
variable path analysis.
Example 1: A Latent Math Proficiency Model
## Warning: lavaan->lav_data_full():
## some observed variances are (at least) a factor 1000
## times larger than others; use varTable(fit) to
## investigate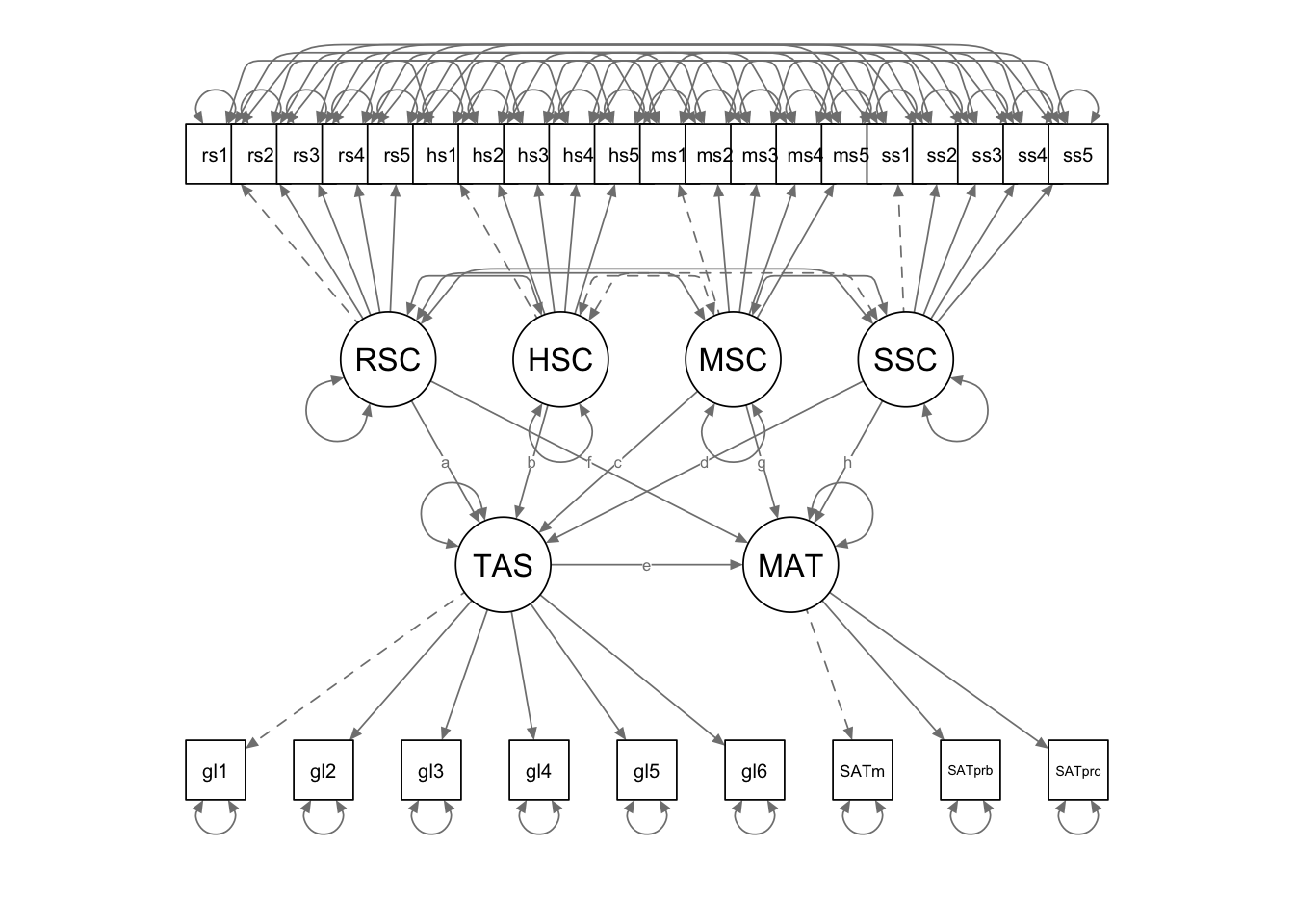
Load Data
library(lavaan)
setwd(mypath)
dat <- read.table("proficiencyraw-female.csv", header = F, sep = ",")
colnames(dat) <- c(paste0("goals",1:6),
paste0("rsc",1:5),
paste0("hsc",1:5),
paste0("msc",1:5),
paste0("ssc",1:5),
"SATvoc",
"SATcomp",
"SATlang",
"SATmath",
"SATprob",
"SATproc")Model Syntax
Here we need to label the path coefficients and define the indirect effects using those labels.
lvpa.Model <- '
# measurement model
RSC =~ rsc1 + rsc2 + rsc3 + rsc4 + rsc5
HSC =~ hsc1 + hsc2 + hsc3 + hsc4 + hsc5
MSC =~ msc1 + msc2 + msc3 + msc4 + msc5
SSC =~ ssc1 + ssc2 + ssc3 + ssc4 + ssc5
TASKGOAL =~ goals1 + goals2 + goals3 + goals4 + goals5 + goals6
MATHPROF =~ SATmath + SATprob + SATproc
HSC ~~ 0*MSC
HSC ~~ 0*SSC
# residual covariances
rsc1~~hsc1+msc1+ssc1
rsc2~~hsc2+msc2+ssc2
rsc3~~hsc3+msc3+ssc3
rsc4~~hsc4+msc4+ssc4
rsc5~~hsc5+msc5+ssc5
hsc1~~msc1+ssc1
hsc2~~msc2+ssc2
hsc3~~msc3+ssc3
hsc4~~msc4+ssc4
hsc5~~msc5+ssc5
msc1~~ssc1
msc2~~ssc2
msc3~~ssc3
msc4~~ssc4
msc5~~ssc5
# structural model for the latent variables
TASKGOAL ~ a*RSC + b*HSC + c*MSC + d*SSC
MATHPROF ~ e*TASKGOAL + f*RSC + g*MSC + h*SSC
# indirect effects
MTHindRSC := e*a
MTHindHSC := e*b
MTHindMSC := e*c
MTHindSSC := e*d
'Fit the model
Fit the model to the data with bootstrapped indirect effects.
lvpa.Fit <- sem(lvpa.Model, data = dat, se = "bootstrap", bootstrap = 5000)Examine the results
You could print out the bootstrapped parameter estimates and
confidence intervals using the parameterEstimates function.
To print out the standardized solutions, you could use the
standardizedSolution function.
summary(lvpa.Fit, fit.measures = T, standardized = T)## lavaan 0.6-19 ended normally after 127 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 100
##
## Number of observations 1000
##
## Model Test User Model:
##
## Test statistic 387.064
## Degrees of freedom 335
## P-value (Chi-square) 0.026
##
## Model Test Baseline Model:
##
## Test statistic 16011.747
## Degrees of freedom 406
## P-value 0.000
##
## User Model versus Baseline Model:
##
## Comparative Fit Index (CFI) 0.997
## Tucker-Lewis Index (TLI) 0.996
##
## Loglikelihood and Information Criteria:
##
## Loglikelihood user model (H0) -51499.857
## Loglikelihood unrestricted model (H1) -51306.326
##
## Akaike (AIC) 103199.715
## Bayesian (BIC) 103690.490
## Sample-size adjusted Bayesian (SABIC) 103372.885
##
## Root Mean Square Error of Approximation:
##
## RMSEA 0.012
## 90 Percent confidence interval - lower 0.005
## 90 Percent confidence interval - upper 0.018
## P-value H_0: RMSEA <= 0.050 1.000
## P-value H_0: RMSEA >= 0.080 0.000
##
## Standardized Root Mean Square Residual:
##
## SRMR 0.032
##
## Parameter Estimates:
##
## Standard errors Standard
## Information Expected
## Information saturated (h1) model Structured
##
## Latent Variables:
## Estimate Std.Err z-value P(>|z|)
## RSC =~
## rsc1 1.000
## rsc2 1.245 0.064 19.435 0.000
## rsc3 1.003 0.059 17.088 0.000
## rsc4 1.205 0.061 19.767 0.000
## rsc5 1.043 0.060 17.480 0.000
## HSC =~
## hsc1 1.000
## hsc2 1.067 0.047 22.721 0.000
## hsc3 0.964 0.048 20.069 0.000
## hsc4 0.880 0.047 18.809 0.000
## hsc5 1.146 0.050 23.008 0.000
## MSC =~
## msc1 1.000
## msc2 0.942 0.032 29.685 0.000
## msc3 0.900 0.032 28.092 0.000
## msc4 0.962 0.034 28.513 0.000
## msc5 0.894 0.032 27.586 0.000
## SSC =~
## ssc1 1.000
## ssc2 0.985 0.029 33.902 0.000
## ssc3 0.820 0.029 28.147 0.000
## ssc4 0.941 0.031 30.051 0.000
## ssc5 0.840 0.031 27.250 0.000
## TASKGOAL =~
## goals1 1.000
## goals2 1.186 0.061 19.370 0.000
## goals3 1.048 0.059 17.808 0.000
## goals4 0.992 0.058 17.216 0.000
## goals5 1.382 0.067 20.566 0.000
## goals6 1.270 0.065 19.613 0.000
## MATHPROF =~
## SATmath 1.000
## SATprob 0.915 0.027 33.719 0.000
## SATproc 1.072 0.034 31.957 0.000
## Std.lv Std.all
##
## 0.820 0.646
## 1.021 0.761
## 0.823 0.644
## 0.988 0.786
## 0.856 0.659
##
## 0.953 0.700
## 1.016 0.803
## 0.919 0.699
## 0.839 0.649
## 1.092 0.820
##
## 1.138 0.817
## 1.072 0.820
## 1.024 0.788
## 1.095 0.797
## 1.017 0.775
##
## 1.168 0.841
## 1.151 0.867
## 0.959 0.766
## 1.100 0.800
## 0.981 0.745
##
## 0.829 0.619
## 0.983 0.767
## 0.868 0.683
## 0.822 0.654
## 1.145 0.840
## 1.052 0.781
##
## 33.631 0.901
## 30.786 0.821
## 36.038 0.795
##
## Regressions:
## Estimate Std.Err z-value P(>|z|)
## TASKGOAL ~
## RSC (a) 0.316 0.043 7.369 0.000
## HSC (b) 0.124 0.033 3.770 0.000
## MSC (c) 0.196 0.032 6.106 0.000
## SSC (d) 0.161 0.030 5.282 0.000
## MATHPROF ~
## TASKGOAL (e) 13.294 1.243 10.695 0.000
## RSC (f) 1.573 1.063 1.481 0.139
## MSC (g) 14.021 0.968 14.478 0.000
## SSC (h) 8.159 0.876 9.312 0.000
## Std.lv Std.all
##
## 0.313 0.313
## 0.143 0.143
## 0.270 0.270
## 0.227 0.227
##
## 0.328 0.328
## 0.038 0.038
## 0.475 0.475
## 0.283 0.283
##
## Covariances:
## Estimate Std.Err z-value P(>|z|)
## HSC ~~
## MSC 0.000
## SSC 0.000
## .rsc1 ~~
## .hsc1 0.121 0.034 3.562 0.000
## .msc1 -0.024 0.029 -0.847 0.397
## .ssc1 0.071 0.028 2.566 0.010
## .rsc2 ~~
## .hsc2 0.122 0.027 4.466 0.000
## .msc2 0.018 0.026 0.706 0.480
## .ssc2 0.036 0.024 1.497 0.134
## .rsc3 ~~
## .hsc3 0.094 0.033 2.837 0.005
## .msc3 0.029 0.028 1.021 0.307
## .ssc3 0.044 0.028 1.574 0.115
## .rsc4 ~~
## .hsc4 0.087 0.030 2.923 0.003
## .msc4 0.015 0.026 0.595 0.552
## .ssc4 0.018 0.026 0.695 0.487
## .rsc5 ~~
## .hsc5 0.096 0.030 3.211 0.001
## .msc5 0.059 0.029 2.004 0.045
## .ssc5 0.120 0.031 3.902 0.000
## .hsc1 ~~
## .msc1 0.043 0.029 1.483 0.138
## .ssc1 0.060 0.028 2.160 0.031
## .hsc2 ~~
## .msc2 0.037 0.023 1.609 0.108
## .ssc2 -0.006 0.021 -0.291 0.771
## .hsc3 ~~
## .msc3 -0.026 0.028 -0.931 0.352
## .ssc3 0.039 0.027 1.443 0.149
## .hsc4 ~~
## .msc4 0.055 0.030 1.846 0.065
## .ssc4 0.097 0.030 3.271 0.001
## .hsc5 ~~
## .msc5 0.018 0.025 0.706 0.480
## .ssc5 0.014 0.026 0.545 0.586
## .msc1 ~~
## .ssc1 0.053 0.024 2.211 0.027
## .msc2 ~~
## .ssc2 0.050 0.020 2.452 0.014
## .msc3 ~~
## .ssc3 0.039 0.024 1.663 0.096
## .msc4 ~~
## .ssc4 0.025 0.026 0.980 0.327
## .msc5 ~~
## .ssc5 0.121 0.027 4.571 0.000
## RSC ~~
## HSC 0.369 0.037 10.076 0.000
## MSC -0.173 0.032 -5.346 0.000
## SSC -0.133 0.032 -4.132 0.000
## MSC ~~
## SSC 0.835 0.059 14.154 0.000
## Std.lv Std.all
##
## 0.000 0.000
## 0.000 0.000
##
## 0.121 0.129
## -0.024 -0.031
## 0.071 0.097
##
## 0.122 0.187
## 0.018 0.028
## 0.036 0.062
##
## 0.094 0.102
## 0.029 0.037
## 0.044 0.056
##
## 0.087 0.113
## 0.015 0.024
## 0.018 0.028
##
## 0.096 0.128
## 0.059 0.073
## 0.120 0.140
##
## 0.043 0.056
## 0.060 0.082
##
## 0.037 0.065
## -0.006 -0.012
##
## -0.026 -0.034
## 0.039 0.052
##
## 0.055 0.067
## 0.097 0.119
##
## 0.018 0.028
## 0.014 0.021
##
## 0.053 0.087
##
## 0.050 0.101
##
## 0.039 0.061
##
## 0.025 0.037
##
## 0.121 0.167
##
## 0.472 0.472
## -0.185 -0.185
## -0.139 -0.139
##
## 0.628 0.628
##
## Variances:
## Estimate Std.Err z-value P(>|z|)
## .rsc1 0.937 0.048 19.421 0.000
## .rsc2 0.758 0.046 16.640 0.000
## .rsc3 0.954 0.049 19.446 0.000
## .rsc4 0.603 0.039 15.626 0.000
## .rsc5 0.954 0.050 19.195 0.000
## .hsc1 0.944 0.049 19.083 0.000
## .hsc2 0.568 0.035 16.056 0.000
## .hsc3 0.883 0.046 19.097 0.000
## .hsc4 0.968 0.049 19.887 0.000
## .hsc5 0.583 0.038 15.275 0.000
## .msc1 0.645 0.036 17.910 0.000
## .msc2 0.561 0.032 17.814 0.000
## .msc3 0.640 0.034 18.726 0.000
## .msc4 0.690 0.037 18.506 0.000
## .msc5 0.686 0.036 19.026 0.000
## .ssc1 0.565 0.033 17.210 0.000
## .ssc2 0.438 0.028 15.916 0.000
## .ssc3 0.649 0.033 19.380 0.000
## .ssc4 0.682 0.037 18.607 0.000
## .ssc5 0.774 0.039 19.751 0.000
## .goals1 1.105 0.053 20.676 0.000
## .goals2 0.677 0.037 18.444 0.000
## .goals3 0.861 0.043 19.979 0.000
## .goals4 0.907 0.045 20.335 0.000
## .goals5 0.548 0.035 15.770 0.000
## .goals6 0.710 0.039 18.067 0.000
## .SATmath 260.998 20.641 12.645 0.000
## .SATprob 459.024 25.912 17.715 0.000
## .SATproc 753.719 40.680 18.528 0.000
## RSC 0.673 0.063 10.728 0.000
## HSC 0.908 0.075 12.054 0.000
## MSC 1.295 0.084 15.332 0.000
## SSC 1.365 0.085 16.093 0.000
## .TASKGOAL 0.473 0.047 10.060 0.000
## .MATHPROF 275.336 23.232 11.852 0.000
## Std.lv Std.all
## 0.937 0.582
## 0.758 0.421
## 0.954 0.585
## 0.603 0.382
## 0.954 0.566
## 0.944 0.510
## 0.568 0.355
## 0.883 0.511
## 0.968 0.579
## 0.583 0.328
## 0.645 0.333
## 0.561 0.328
## 0.640 0.379
## 0.690 0.365
## 0.686 0.399
## 0.565 0.293
## 0.438 0.248
## 0.649 0.414
## 0.682 0.361
## 0.774 0.446
## 1.105 0.617
## 0.677 0.412
## 0.861 0.533
## 0.907 0.573
## 0.548 0.295
## 0.710 0.391
## 260.998 0.187
## 459.024 0.326
## 753.719 0.367
## 1.000 1.000
## 1.000 1.000
## 1.000 1.000
## 1.000 1.000
## 0.689 0.689
## 0.243 0.243
##
## Defined Parameters:
## Estimate Std.Err z-value P(>|z|)
## MTHindRSC 4.205 0.641 6.564 0.000
## MTHindHSC 1.651 0.455 3.628 0.000
## MTHindMSC 2.612 0.456 5.727 0.000
## MTHindSSC 2.139 0.434 4.922 0.000
## Std.lv Std.all
## 0.103 0.103
## 0.047 0.047
## 0.088 0.088
## 0.074 0.074# parameterEstimates(lvpa.Fit, ci = T, standardized = T)
standardizedSolution(lvpa.Fit)## lhs op rhs label est.std se z
## 1 RSC =~ rsc1 0.646 0.022 29.983
## 2 RSC =~ rsc2 0.761 0.017 44.200
## 3 RSC =~ rsc3 0.644 0.022 29.702
## 4 RSC =~ rsc4 0.786 0.016 48.085
## 5 RSC =~ rsc5 0.659 0.021 31.289
## 6 HSC =~ hsc1 0.700 0.019 37.143
## 7 HSC =~ hsc2 0.803 0.015 54.680
## 8 HSC =~ hsc3 0.699 0.019 36.921
## 9 HSC =~ hsc4 0.649 0.021 31.120
## 10 HSC =~ hsc5 0.820 0.014 58.165
## 11 MSC =~ msc1 0.817 0.013 65.321
## 12 MSC =~ msc2 0.820 0.012 66.209
## 13 MSC =~ msc3 0.788 0.014 56.858
## 14 MSC =~ msc4 0.797 0.013 59.152
## 15 MSC =~ msc5 0.775 0.014 53.801
## 16 SSC =~ ssc1 0.841 0.011 74.764
## 17 SSC =~ ssc2 0.867 0.010 86.149
## 18 SSC =~ ssc3 0.766 0.015 51.730
## 19 SSC =~ ssc4 0.800 0.013 60.533
## 20 SSC =~ ssc5 0.745 0.016 47.487
## 21 TASKGOAL =~ goals1 0.619 0.022 28.770
## 22 TASKGOAL =~ goals2 0.767 0.015 49.817
## 23 TASKGOAL =~ goals3 0.683 0.019 35.993
## 24 TASKGOAL =~ goals4 0.654 0.020 32.392
## 25 TASKGOAL =~ goals5 0.840 0.012 68.991
## 26 TASKGOAL =~ goals6 0.781 0.015 52.847
## 27 MATHPROF =~ SATmath 0.901 0.009 100.167
## 28 MATHPROF =~ SATprob 0.821 0.012 66.390
## 29 MATHPROF =~ SATproc 0.795 0.014 58.814
## 30 HSC ~~ MSC 0.000 0.000 NA
## 31 HSC ~~ SSC 0.000 0.000 NA
## 32 rsc1 ~~ hsc1 0.129 0.035 3.663
## 33 rsc1 ~~ msc1 -0.031 0.037 -0.848
## 34 rsc1 ~~ ssc1 0.097 0.037 2.596
## 35 rsc2 ~~ hsc2 0.187 0.040 4.717
## 36 rsc2 ~~ msc2 0.028 0.040 0.706
## 37 rsc2 ~~ ssc2 0.062 0.041 1.504
## 38 rsc3 ~~ hsc3 0.102 0.035 2.891
## 39 rsc3 ~~ msc3 0.037 0.036 1.022
## 40 rsc3 ~~ ssc3 0.056 0.036 1.581
## 41 rsc4 ~~ hsc4 0.113 0.038 2.991
## 42 rsc4 ~~ msc4 0.024 0.040 0.595
## 43 rsc4 ~~ ssc4 0.028 0.040 0.696
## 44 rsc5 ~~ hsc5 0.128 0.039 3.306
## 45 rsc5 ~~ msc5 0.073 0.036 2.015
## 46 rsc5 ~~ ssc5 0.140 0.035 4.002
## 47 hsc1 ~~ msc1 0.056 0.037 1.489
## 48 hsc1 ~~ ssc1 0.082 0.038 2.178
## 49 hsc2 ~~ msc2 0.065 0.040 1.617
## 50 hsc2 ~~ ssc2 -0.012 0.042 -0.291
## 51 hsc3 ~~ msc3 -0.034 0.037 -0.931
## 52 hsc3 ~~ ssc3 0.052 0.036 1.449
## 53 hsc4 ~~ msc4 0.067 0.036 1.857
## 54 hsc4 ~~ ssc4 0.119 0.036 3.331
## 55 hsc5 ~~ msc5 0.028 0.040 0.707
## 56 hsc5 ~~ ssc5 0.021 0.039 0.545
## 57 msc1 ~~ ssc1 0.087 0.039 2.241
## 58 msc2 ~~ ssc2 0.101 0.040 2.493
## 59 msc3 ~~ ssc3 0.061 0.036 1.674
## 60 msc4 ~~ ssc4 0.037 0.037 0.983
## 61 msc5 ~~ ssc5 0.167 0.035 4.757
## 62 TASKGOAL ~ RSC a 0.313 0.037 8.355
## 63 TASKGOAL ~ HSC b 0.143 0.037 3.877
## 64 TASKGOAL ~ MSC c 0.270 0.042 6.473
## 65 TASKGOAL ~ SSC d 0.227 0.041 5.510
## 66 MATHPROF ~ TASKGOAL e 0.328 0.027 12.139
## 67 MATHPROF ~ RSC f 0.038 0.026 1.484
## 68 MATHPROF ~ MSC g 0.475 0.029 16.149
## 69 MATHPROF ~ SSC h 0.283 0.029 9.632
## 70 rsc1 ~~ rsc1 0.582 0.028 20.892
## 71 rsc2 ~~ rsc2 0.421 0.026 16.060
## 72 rsc3 ~~ rsc3 0.585 0.028 20.913
## 73 rsc4 ~~ rsc4 0.382 0.026 14.842
## 74 rsc5 ~~ rsc5 0.566 0.028 20.383
## 75 hsc1 ~~ hsc1 0.510 0.026 19.314
## 76 hsc2 ~~ hsc2 0.355 0.024 15.047
## 77 hsc3 ~~ hsc3 0.511 0.026 19.313
## 78 hsc4 ~~ hsc4 0.579 0.027 21.399
## 79 hsc5 ~~ hsc5 0.328 0.023 14.215
## 80 msc1 ~~ msc1 0.333 0.020 16.272
## 81 msc2 ~~ msc2 0.328 0.020 16.162
## 82 msc3 ~~ msc3 0.379 0.022 17.326
## 83 msc4 ~~ msc4 0.365 0.021 17.019
## 84 msc5 ~~ msc5 0.399 0.022 17.831
## 85 ssc1 ~~ ssc1 0.293 0.019 15.459
## 86 ssc2 ~~ ssc2 0.248 0.017 14.223
## 87 ssc3 ~~ ssc3 0.414 0.023 18.263
## 88 ssc4 ~~ ssc4 0.361 0.021 17.082
## 89 ssc5 ~~ ssc5 0.446 0.023 19.090
## 90 goals1 ~~ goals1 0.617 0.027 23.149
## 91 goals2 ~~ goals2 0.412 0.024 17.471
## 92 goals3 ~~ goals3 0.533 0.026 20.574
## 93 goals4 ~~ goals4 0.573 0.026 21.723
## 94 goals5 ~~ goals5 0.295 0.020 14.401
## 95 goals6 ~~ goals6 0.391 0.023 16.939
## 96 SATmath ~~ SATmath 0.187 0.016 11.557
## 97 SATprob ~~ SATprob 0.326 0.020 16.077
## 98 SATproc ~~ SATproc 0.367 0.022 17.067
## 99 RSC ~~ RSC 1.000 0.000 NA
## 100 HSC ~~ HSC 1.000 0.000 NA
## pvalue ci.lower ci.upper
## 1 0.000 0.604 0.689
## 2 0.000 0.727 0.795
## 3 0.000 0.602 0.687
## 4 0.000 0.754 0.818
## 5 0.000 0.618 0.700
## 6 0.000 0.663 0.737
## 7 0.000 0.774 0.832
## 8 0.000 0.662 0.736
## 9 0.000 0.608 0.690
## 10 0.000 0.792 0.847
## 11 0.000 0.792 0.841
## 12 0.000 0.795 0.844
## 13 0.000 0.761 0.815
## 14 0.000 0.770 0.823
## 15 0.000 0.747 0.804
## 16 0.000 0.819 0.863
## 17 0.000 0.847 0.887
## 18 0.000 0.737 0.795
## 19 0.000 0.774 0.825
## 20 0.000 0.714 0.775
## 21 0.000 0.577 0.661
## 22 0.000 0.736 0.797
## 23 0.000 0.646 0.720
## 24 0.000 0.614 0.693
## 25 0.000 0.816 0.864
## 26 0.000 0.752 0.810
## 27 0.000 0.884 0.919
## 28 0.000 0.797 0.845
## 29 0.000 0.769 0.822
## 30 NA 0.000 0.000
## 31 NA 0.000 0.000
## 32 0.000 0.060 0.198
## 33 0.396 -0.104 0.041
## 34 0.009 0.024 0.170
## 35 0.000 0.109 0.264
## 36 0.480 -0.050 0.105
## 37 0.133 -0.019 0.143
## 38 0.004 0.033 0.172
## 39 0.307 -0.034 0.108
## 40 0.114 -0.014 0.126
## 41 0.003 0.039 0.188
## 42 0.552 -0.054 0.102
## 43 0.486 -0.050 0.105
## 44 0.001 0.052 0.204
## 45 0.044 0.002 0.144
## 46 0.000 0.072 0.209
## 47 0.136 -0.018 0.129
## 48 0.029 0.008 0.156
## 49 0.106 -0.014 0.143
## 50 0.771 -0.095 0.070
## 51 0.352 -0.106 0.038
## 52 0.147 -0.018 0.123
## 53 0.063 -0.004 0.138
## 54 0.001 0.049 0.189
## 55 0.479 -0.050 0.106
## 56 0.585 -0.055 0.098
## 57 0.025 0.011 0.164
## 58 0.013 0.022 0.180
## 59 0.094 -0.010 0.132
## 60 0.326 -0.037 0.110
## 61 0.000 0.098 0.235
## 62 0.000 0.240 0.387
## 63 0.000 0.071 0.215
## 64 0.000 0.188 0.352
## 65 0.000 0.146 0.308
## 66 0.000 0.275 0.380
## 67 0.138 -0.012 0.089
## 68 0.000 0.417 0.532
## 69 0.000 0.226 0.341
## 70 0.000 0.528 0.637
## 71 0.000 0.369 0.472
## 72 0.000 0.530 0.640
## 73 0.000 0.331 0.432
## 74 0.000 0.511 0.620
## 75 0.000 0.458 0.562
## 76 0.000 0.309 0.401
## 77 0.000 0.459 0.563
## 78 0.000 0.526 0.632
## 79 0.000 0.283 0.374
## 80 0.000 0.292 0.373
## 81 0.000 0.288 0.368
## 82 0.000 0.336 0.422
## 83 0.000 0.323 0.407
## 84 0.000 0.355 0.442
## 85 0.000 0.255 0.330
## 86 0.000 0.214 0.282
## 87 0.000 0.369 0.458
## 88 0.000 0.319 0.402
## 89 0.000 0.400 0.491
## 90 0.000 0.565 0.669
## 91 0.000 0.366 0.459
## 92 0.000 0.483 0.584
## 93 0.000 0.521 0.625
## 94 0.000 0.254 0.335
## 95 0.000 0.345 0.436
## 96 0.000 0.156 0.219
## 97 0.000 0.287 0.366
## 98 0.000 0.325 0.409
## 99 NA 1.000 1.000
## 100 NA 1.000 1.000
## [ reached 'max' / getOption("max.print") -- omitted 12 rows ]Example 2: Two-step LVPA
The example corresponds to the “exercise behavior example” in your course slides. Please refer to the slides for more information about the data.
Read in the data
# lower half of the correlation matrix
cormat <- '
1.000
.812 1.000
.819 .752 1.000
.334 .344 .228 1.000
.177 .094 .141 .363 1.000
.363 .383 .387 .241 .273 1.000
.239 .258 .275 .286 .389 .445 1.000
.243 .293 .234 .116 .096 .222 .344 1.000
.672 .616 .621 .277 .137 .458 .315 .246 1.000
.464 .620 .514 .213 .173 .430 .387 .132 .680 1.000
.612 .640 .719 .192 .090 .509 .336 .230 .819 .676 1.000
.331 .391 .310 .435 .263 .409 .298 .256 .446 .395 .411 1.000
.209 .214 .286 .319 .671 .423 .334 .246 .308 .268 .280 .573 1.000
.298 .358 .361 .171 .232 .791 .286 .057 .433 .387 .477 .389 .445 1.000
.309 .303 .381 .132 .307 .637 .459 .267 .468 .406 .458 .554 .514 .551 1.000
.056 .086 .092 .090 .201 .123 .247 .403 .176 .076 .131 .318 .213 .056 .342 1.000
'
# standard deviations
sdev <- c(2.46, 1.76, 2.74, 2.04, 2.13, 4.30, 1.90, 1.90, 2.63, 1.89, 2.84, 2.34, 2.27, 4.86, 2.66, 1.94)
# compute the variance-covariance matrix
Cmat <- getCov(cormat)
Dmat <- diag(sdev)
covmat <- Dmat %*% Cmat %*% Dmat
# assign variable names
rownames(covmat) <- colnames(covmat) <- c('ss1t1', 'ss2t1', 'ss3t1', 'se1t1', 'se2t1', 'eb1t1', 'eb2t1',
'eb3t1','ss1t2', 'ss2t2', 'ss3t2', 'se1t2', 'se2t2', 'eb1t2',
'eb2t2', 'eb3t2')Step 1: Measurement phase
Fit the initial measurement model
We first fit a model where all latent factors are allowed to covary. In the following model syntax:
We only specified six formulas, each defining a latent factor. By default, all exogenous latent variables are correlated. We thus do not need to manually covary them (but it does no harm if you wish to be more explicit about the model you fit by writing down all the formulas).
We had constrained measurement invariance across time points by imposing equality constraints on the factor loadings. To constrain model parameters to be equal, we need to use the same label for the corresponding parameters. To label a parameter, we just multiply it with a character string (e.g., a, b, c…).
DD.model <- '
SOCSUP1 =~ ss1t1 + a*ss2t1 + b*ss3t1
SELFEFF1 =~ se1t1 + c*se2t1
EXBEH1 =~ eb1t1 + d*eb2t1 + e*eb3t1
SOCSUP2 =~ ss1t2 + a*ss2t2 + b*ss3t2
SELFEFF2 =~ se1t2 + c*se2t2
EXBEH2 =~ eb1t2 + d*eb2t2 + e*eb3t2
'
DD.fit <- sem(DD.model, sample.cov = covmat, sample.nobs = 84)
summary(DD.fit, fit.measures = T, standardized = T)## lavaan 0.6-19 ended normally after 143 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 47
## Number of equality constraints 5
##
## Number of observations 84
##
## Model Test User Model:
##
## Test statistic 194.043
## Degrees of freedom 94
## P-value (Chi-square) 0.000
##
## Model Test Baseline Model:
##
## Test statistic 904.195
## Degrees of freedom 120
## P-value 0.000
##
## User Model versus Baseline Model:
##
## Comparative Fit Index (CFI) 0.872
## Tucker-Lewis Index (TLI) 0.837
##
## Loglikelihood and Information Criteria:
##
## Loglikelihood user model (H0) -2738.760
## Loglikelihood unrestricted model (H1) -2641.739
##
## Akaike (AIC) 5561.520
## Bayesian (BIC) 5663.614
## Sample-size adjusted Bayesian (SABIC) 5531.124
##
## Root Mean Square Error of Approximation:
##
## RMSEA 0.113
## 90 Percent confidence interval - lower 0.090
## 90 Percent confidence interval - upper 0.135
## P-value H_0: RMSEA <= 0.050 0.000
## P-value H_0: RMSEA >= 0.080 0.990
##
## Standardized Root Mean Square Residual:
##
## SRMR 0.104
##
## Parameter Estimates:
##
## Standard errors Standard
## Information Expected
## Information saturated (h1) model Structured
##
## Latent Variables:
## Estimate Std.Err z-value P(>|z|)
## SOCSUP1 =~
## ss1t1 1.000
## ss2t1 (a) 0.648 0.044 14.686 0.000
## ss3t1 (b) 1.087 0.063 17.366 0.000
## SELFEFF1 =~
## se1t1 1.000
## se2t1 (c) 1.382 0.190 7.281 0.000
## EXBEH1 =~
## eb1t1 1.000
## eb2t1 (d) 0.382 0.043 8.976 0.000
## eb3t1 (e) 0.117 0.039 3.013 0.003
## SOCSUP2 =~
## ss1t2 1.000
## ss2t2 (a) 0.648 0.044 14.686 0.000
## ss3t2 (b) 1.087 0.063 17.366 0.000
## SELFEFF2 =~
## se1t2 1.000
## se2t2 (c) 1.382 0.190 7.281 0.000
## EXBEH2 =~
## eb1t2 1.000
## eb2t2 (d) 0.382 0.043 8.976 0.000
## eb3t2 (e) 0.117 0.039 3.013 0.003
## Std.lv Std.all
##
## 2.265 0.923
## 1.468 0.860
## 2.463 0.890
##
## 1.087 0.525
## 1.503 0.719
##
## 3.485 0.851
## 1.332 0.629
## 0.409 0.218
##
## 2.330 0.896
## 1.511 0.772
## 2.534 0.909
##
## 1.451 0.634
## 2.006 0.884
##
## 4.025 0.778
## 1.538 0.637
## 0.472 0.244
##
## Covariances:
## Estimate Std.Err z-value P(>|z|)
## SOCSUP1 ~~
## SELFEFF1 0.823 0.384 2.144 0.032
## EXBEH1 3.966 1.118 3.547 0.000
## SOCSUP2 4.185 0.814 5.139 0.000
## SELFEFF2 1.125 0.452 2.487 0.013
## EXBEH2 4.529 1.350 3.354 0.001
## SELFEFF1 ~~
## EXBEH1 2.088 0.689 3.032 0.002
## SOCSUP2 0.684 0.390 1.753 0.080
## SELFEFF2 1.469 0.435 3.379 0.001
## EXBEH2 2.137 0.813 2.629 0.009
## EXBEH1 ~~
## SOCSUP2 5.256 1.221 4.304 0.000
## SELFEFF2 3.123 0.861 3.629 0.000
## EXBEH2 15.459 2.835 5.452 0.000
## SOCSUP2 ~~
## SELFEFF2 1.463 0.491 2.981 0.003
## EXBEH2 6.491 1.500 4.328 0.000
## SELFEFF2 ~~
## EXBEH2 4.526 1.125 4.024 0.000
## Std.lv Std.all
##
## 0.334 0.334
## 0.502 0.502
## 0.793 0.793
## 0.342 0.342
## 0.497 0.497
##
## 0.551 0.551
## 0.270 0.270
## 0.931 0.931
## 0.488 0.488
##
## 0.647 0.647
## 0.618 0.618
## 1.102 1.102
##
## 0.433 0.433
## 0.692 0.692
##
## 0.775 0.775
##
## Variances:
## Estimate Std.Err z-value P(>|z|)
## .ss1t1 0.892 0.242 3.685 0.000
## .ss2t1 0.757 0.149 5.096 0.000
## .ss3t1 1.585 0.345 4.591 0.000
## .se1t1 3.100 0.532 5.826 0.000
## .se2t1 2.107 0.551 3.821 0.000
## .eb1t1 4.625 1.350 3.426 0.001
## .eb2t1 2.702 0.451 5.996 0.000
## .eb3t1 3.358 0.520 6.453 0.000
## .ss1t2 1.341 0.314 4.272 0.000
## .ss2t2 1.547 0.270 5.727 0.000
## .ss3t2 1.349 0.345 3.905 0.000
## .se1t2 3.132 0.537 5.838 0.000
## .se2t2 1.130 0.478 2.364 0.018
## .eb1t2 10.557 2.241 4.710 0.000
## .eb2t2 3.463 0.579 5.983 0.000
## .eb3t2 3.540 0.545 6.491 0.000
## SOCSUP1 5.130 0.906 5.664 0.000
## SELFEFF1 1.182 0.415 2.847 0.004
## EXBEH1 12.148 2.663 4.562 0.000
## SOCSUP2 5.431 0.984 5.521 0.000
## SELFEFF2 2.106 0.613 3.434 0.001
## EXBEH2 16.198 3.946 4.105 0.000
## Std.lv Std.all
## 0.892 0.148
## 0.757 0.260
## 1.585 0.207
## 3.100 0.724
## 2.107 0.483
## 4.625 0.276
## 2.702 0.604
## 3.358 0.953
## 1.341 0.198
## 1.547 0.404
## 1.349 0.174
## 3.132 0.598
## 1.130 0.219
## 10.557 0.395
## 3.463 0.594
## 3.540 0.941
## 1.000 1.000
## 1.000 1.000
## 1.000 1.000
## 1.000 1.000
## 1.000 1.000
## 1.000 1.000Examine model fit and modification indicies
The initial measurement model fit poorly with the data.
Modification indices can be obtained by calling the function
modindices(). You can ask R to only print out the
modification indices that are only greater than 3.841 (the critical
value of \(\chi^2\) statistic with
\(df=1\) and \(p=0.05\).)
DD.mod <- modindices(DD.fit)
with(DD.mod, subset(DD.mod[order(mi, decreasing = T), ], subset = mi >= 3.84))| lhs | op | rhs | mi | epc | sepc.lv | sepc.all | sepc.nox | |
|---|---|---|---|---|---|---|---|---|
| 211 | eb1t1 | ~~ | eb1t2 | 29.554874 | 7.0285637 | 7.0285637 | 1.0058475 | 1.0058475 |
| 200 | se2t1 | ~~ | se2t2 | 21.492411 | 2.2630455 | 2.2630455 | 1.4665175 | 1.4665175 |
| 199 | se2t1 | ~~ | se1t2 | 14.130701 | -1.5009328 | -1.5009328 | -0.5841848 | -0.5841848 |
| 175 | ss3t1 | ~~ | ss3t2 | 12.718190 | 0.8439916 | 0.8439916 | 0.5771916 | 0.5771916 |
| 95 | EXBEH1 | =~ | se2t2 | 12.183893 | -0.4680217 | -1.6312398 | -0.7185481 | -0.7185481 |
| 146 | ss1t1 | ~~ | ss1t2 | 11.838854 | 0.6514608 | 0.6514608 | 0.5958094 | 0.5958094 |
| 138 | EXBEH2 | =~ | se2t2 | 10.772413 | -0.3665133 | -1.4750935 | -0.6497669 | -0.6497669 |
| 204 | eb1t1 | ~~ | eb2t1 | 10.652358 | -3.6638068 | -3.6638068 | -1.0364330 | -1.0364330 |
| 230 | eb3t1 | ~~ | eb3t2 | 10.499260 | 1.2293109 | 1.2293109 | 0.3565765 | 0.3565765 |
| 161 | ss2t1 | ~~ | ss2t2 | 10.267321 | 0.4479934 | 0.4479934 | 0.4138389 | 0.4138389 |
| 97 | EXBEH1 | =~ | eb2t2 | 10.257542 | 0.2954189 | 1.0296509 | 0.4265327 | 0.4265327 |
| 108 | SOCSUP2 | =~ | se2t2 | 9.100605 | -0.4337218 | -1.0107596 | -0.4452315 | -0.4452315 |
| 228 | eb3t1 | ~~ | eb1t2 | 8.742146 | -2.1606781 | -2.1606781 | -0.3629074 | -0.3629074 |
| 107 | SOCSUP2 | =~ | se1t2 | 8.550785 | 0.3020341 | 0.7038702 | 0.3075451 | 0.3075451 |
| 188 | se1t1 | ~~ | se1t2 | 8.350035 | 1.1698638 | 1.1698638 | 0.3754016 | 0.3754016 |
| 189 | se1t1 | ~~ | se2t2 | 8.130852 | -1.0429171 | -1.0429171 | -0.5572058 | -0.5572058 |
| 132 | EXBEH2 | =~ | eb2t1 | 7.155722 | -0.1822889 | -0.7336519 | -0.3468041 | -0.3468041 |
| 123 | SELFEFF2 | =~ | eb1t2 | 7.135689 | -1.1561905 | -1.6777238 | -0.3243521 | -0.3243521 |
| 124 | SELFEFF2 | =~ | eb2t2 | 7.087618 | 0.5394738 | 0.7828191 | 0.3242827 | 0.3242827 |
| 148 | ss1t1 | ~~ | ss3t2 | 6.705066 | -0.5101159 | -0.5101159 | -0.4651139 | -0.4651139 |
| 173 | ss3t1 | ~~ | ss1t2 | 6.483371 | -0.5722017 | -0.5722017 | -0.3925179 | -0.3925179 |
| 84 | SELFEFF1 | =~ | eb2t2 | 5.902730 | 0.6231179 | 0.6775664 | 0.2806818 | 0.2806818 |
| 131 | EXBEH2 | =~ | eb1t1 | 5.901371 | 0.3411144 | 1.3728713 | 0.3352153 | 0.3352153 |
| 68 | SOCSUP1 | =~ | se2t2 | 5.426477 | -0.3316135 | -0.7510658 | -0.3308385 | -0.3308385 |
| 137 | EXBEH2 | =~ | se1t2 | 5.414126 | 0.1757127 | 0.7071847 | 0.3089933 | 0.3089933 |
| 67 | SOCSUP1 | =~ | se1t2 | 5.333676 | 0.2364748 | 0.5355878 | 0.2340167 | 0.2340167 |
| 147 | ss1t1 | ~~ | ss2t2 | 5.129417 | -0.3965698 | -0.3965698 | -0.3376218 | -0.3376218 |
| 220 | eb2t1 | ~~ | eb1t2 | 5.121894 | -1.6245688 | -1.6245688 | -0.3041807 | -0.3041807 |
| 257 | eb1t2 | ~~ | eb3t2 | 4.889472 | -1.7042414 | -1.7042414 | -0.2787833 | -0.2787833 |
| 258 | eb2t2 | ~~ | eb3t2 | 4.840989 | 0.8922979 | 0.8922979 | 0.2548602 | 0.2548602 |
| 251 | se1t2 | ~~ | eb2t2 | 4.517625 | 0.8641458 | 0.8641458 | 0.2623798 | 0.2623798 |
| 194 | se2t1 | ~~ | eb2t1 | 4.201917 | 0.6461529 | 0.6461529 | 0.2707889 | 0.2707889 |
| 14 | EXBEH2 | =~ | eb1t2 | 4.175087 | -0.3765547 | -1.5155068 | -0.2929909 | -0.2929909 |
| 177 | ss3t1 | ~~ | se2t2 | 4.114356 | 0.4796058 | 0.4796058 | 0.3583504 | 0.3583504 |
| 214 | eb2t1 | ~~ | eb3t1 | 3.900313 | 0.6926547 | 0.6926547 | 0.2299671 | 0.2299671 |
| 163 | ss2t1 | ~~ | se1t2 | 3.875494 | 0.3858542 | 0.3858542 | 0.2504952 | 0.2504952 |
Fit a modified measurement model
With the modification indices and the substantive theory, we modified the initial measurement model and fitted it to the data. In the following model syntax, we allowed the residuals to covary between the same item at different time points. We then check the model fit and modification indicies for this modified model.
DD.model2 <- '
SOCSUP1 =~ ss1t1 + a*ss2t1 + b*ss3t1
SELFEFF1 =~ se1t1 + c*se2t1
EXBEH1 =~ eb1t1 + d*eb2t1 + e*eb3t1
SOCSUP2 =~ ss1t2 + a*ss2t2 + b*ss3t2
SELFEFF2 =~ se1t2 + c*se2t2
EXBEH2 =~ eb1t2 + d*eb2t2 + e*eb3t2
ss1t1~~ss1t2
ss2t1~~ss2t2
ss3t1~~ss3t2
se1t1~~se1t2
se2t1~~se2t2
eb1t1~~eb1t2
eb2t1~~eb2t2
eb3t1~~eb3t2
'
DD.fit2 <- sem(DD.model2, sample.cov = covmat, sample.nobs = 84)
summary(DD.fit2, fit.measures = T, standardized = T, modindices = T)## lavaan 0.6-19 ended normally after 171 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 55
## Number of equality constraints 5
##
## Number of observations 84
##
## Model Test User Model:
##
## Test statistic 105.692
## Degrees of freedom 86
## P-value (Chi-square) 0.074
##
## Model Test Baseline Model:
##
## Test statistic 904.195
## Degrees of freedom 120
## P-value 0.000
##
## User Model versus Baseline Model:
##
## Comparative Fit Index (CFI) 0.975
## Tucker-Lewis Index (TLI) 0.965
##
## Loglikelihood and Information Criteria:
##
## Loglikelihood user model (H0) -2694.584
## Loglikelihood unrestricted model (H1) -2641.739
##
## Akaike (AIC) 5489.169
## Bayesian (BIC) 5610.710
## Sample-size adjusted Bayesian (SABIC) 5452.983
##
## Root Mean Square Error of Approximation:
##
## RMSEA 0.052
## 90 Percent confidence interval - lower 0.000
## 90 Percent confidence interval - upper 0.083
## P-value H_0: RMSEA <= 0.050 0.439
## P-value H_0: RMSEA >= 0.080 0.072
##
## Standardized Root Mean Square Residual:
##
## SRMR 0.080
##
## Parameter Estimates:
##
## Standard errors Standard
## Information Expected
## Information saturated (h1) model Structured
##
## Latent Variables:
## Estimate Std.Err z-value P(>|z|)
## SOCSUP1 =~
## ss1t1 1.000
## ss2t1 (a) 0.633 0.048 13.291 0.000
## ss3t1 (b) 1.028 0.070 14.654 0.000
## SELFEFF1 =~
## se1t1 1.000
## se2t1 (c) 0.822 0.140 5.849 0.000
## EXBEH1 =~
## eb1t1 1.000
## eb2t1 (d) 0.543 0.086 6.311 0.000
## eb3t1 (e) 0.224 0.062 3.638 0.000
## SOCSUP2 =~
## ss1t2 1.000
## ss2t2 (a) 0.633 0.048 13.291 0.000
## ss3t2 (b) 1.028 0.070 14.654 0.000
## SELFEFF2 =~
## se1t2 1.000
## se2t2 (c) 0.822 0.140 5.849 0.000
## EXBEH2 =~
## eb1t2 1.000
## eb2t2 (d) 0.543 0.086 6.311 0.000
## eb3t2 (e) 0.224 0.062 3.638 0.000
## Std.lv Std.all
##
## 2.322 0.938
## 1.469 0.859
## 2.387 0.875
##
## 1.312 0.640
## 1.078 0.519
##
## 2.953 0.708
## 1.604 0.764
## 0.661 0.354
##
## 2.396 0.908
## 1.516 0.776
## 2.463 0.894
##
## 1.908 0.818
## 1.567 0.704
##
## 3.212 0.644
## 1.745 0.728
## 0.719 0.370
##
## Covariances:
## Estimate Std.Err z-value P(>|z|)
## .ss1t1 ~~
## .ss1t2 0.444 0.227 1.959 0.050
## .ss2t1 ~~
## .ss2t2 0.392 0.153 2.569 0.010
## .ss3t1 ~~
## .ss3t2 0.696 0.288 2.416 0.016
## .se1t1 ~~
## .se1t2 0.628 0.445 1.409 0.159
## .se2t1 ~~
## .se2t2 1.916 0.448 4.274 0.000
## .eb1t1 ~~
## .eb1t2 7.510 1.862 4.034 0.000
## .eb2t1 ~~
## .eb2t2 -0.281 0.399 -0.704 0.481
## .eb3t1 ~~
## .eb3t2 0.988 0.373 2.648 0.008
## SOCSUP1 ~~
## SELFEFF1 1.402 0.502 2.794 0.005
## EXBEH1 3.267 1.030 3.173 0.002
## SOCSUP2 4.157 0.855 4.859 0.000
## SELFEFF2 1.917 0.623 3.077 0.002
## EXBEH2 3.411 1.159 2.943 0.003
## SELFEFF1 ~~
## EXBEH1 2.461 0.771 3.194 0.001
## SOCSUP2 1.199 0.513 2.339 0.019
## SELFEFF2 1.590 0.571 2.784 0.005
## EXBEH2 2.234 0.845 2.643 0.008
## EXBEH1 ~~
## SOCSUP2 4.380 1.166 3.756 0.000
## SELFEFF2 3.756 1.034 3.635 0.000
## EXBEH2 8.896 2.665 3.339 0.001
## SOCSUP2 ~~
## SELFEFF2 2.532 0.682 3.714 0.000
## EXBEH2 5.147 1.354 3.801 0.000
## SELFEFF2 ~~
## EXBEH2 5.159 1.269 4.066 0.000
## Std.lv Std.all
##
## 0.444 0.469
##
## 0.392 0.363
##
## 0.696 0.426
##
## 0.628 0.298
##
## 1.916 0.682
##
## 7.510 0.667
##
## -0.281 -0.126
##
## 0.988 0.313
##
## 0.460 0.460
## 0.477 0.477
## 0.747 0.747
## 0.433 0.433
## 0.457 0.457
##
## 0.636 0.636
## 0.382 0.382
## 0.635 0.635
## 0.530 0.530
##
## 0.619 0.619
## 0.667 0.667
## 0.938 0.938
##
## 0.554 0.554
## 0.669 0.669
##
## 0.842 0.842
##
## Variances:
## Estimate Std.Err z-value P(>|z|)
## .ss1t1 0.736 0.263 2.795 0.005
## .ss2t1 0.767 0.154 4.974 0.000
## .ss3t1 1.749 0.374 4.682 0.000
## .se1t1 2.475 0.637 3.886 0.000
## .se2t1 3.158 0.594 5.321 0.000
## .eb1t1 8.697 1.808 4.811 0.000
## .eb2t1 1.835 0.472 3.889 0.000
## .eb3t1 3.046 0.485 6.283 0.000
## .ss1t2 1.216 0.346 3.516 0.000
## .ss2t2 1.519 0.271 5.595 0.000
## .ss3t2 1.527 0.389 3.924 0.000
## .se1t2 1.795 0.583 3.078 0.002
## .se2t2 2.502 0.523 4.782 0.000
## .eb1t2 14.583 2.625 5.556 0.000
## .eb2t2 2.694 0.613 4.392 0.000
## .eb3t2 3.269 0.518 6.317 0.000
## SOCSUP1 5.392 0.952 5.663 0.000
## SELFEFF1 1.720 0.624 2.758 0.006
## EXBEH1 8.717 2.538 3.435 0.001
## SOCSUP2 5.740 1.047 5.483 0.000
## SELFEFF2 3.639 0.932 3.905 0.000
## EXBEH2 10.316 3.297 3.129 0.002
## Std.lv Std.all
## 0.736 0.120
## 0.767 0.262
## 1.749 0.235
## 2.475 0.590
## 3.158 0.731
## 8.697 0.499
## 1.835 0.416
## 3.046 0.874
## 1.216 0.175
## 1.519 0.398
## 1.527 0.201
## 1.795 0.330
## 2.502 0.505
## 14.583 0.586
## 2.694 0.469
## 3.269 0.863
## 1.000 1.000
## 1.000 1.000
## 1.000 1.000
## 1.000 1.000
## 1.000 1.000
## 1.000 1.000
##
## Modification Indices:
##
## lhs op rhs mi epc sepc.lv sepc.all
## 1 SOCSUP1 =~ ss1t1 0.372 -0.049 -0.115 -0.046
## 2 SELFEFF1 =~ se1t1 0.062 -0.071 -0.093 -0.046
## 3 EXBEH1 =~ eb1t1 8.987 0.581 1.715 0.411
## 4 SOCSUP2 =~ ss1t2 0.372 0.049 0.119 0.045
## 5 SELFEFF2 =~ se1t2 0.062 0.071 0.136 0.058
## 6 EXBEH2 =~ eb1t2 8.987 -0.581 -1.866 -0.374
## 7 SOCSUP1 =~ se1t1 0.325 0.062 0.144 0.070
## 8 SOCSUP1 =~ se2t1 0.325 -0.051 -0.118 -0.057
## 9 SOCSUP1 =~ eb1t1 2.494 0.290 0.673 0.161
## 10 SOCSUP1 =~ eb2t1 5.503 -0.240 -0.557 -0.265
## 11 SOCSUP1 =~ eb3t1 2.230 0.130 0.301 0.161
## 12 SOCSUP1 =~ ss1t2 0.014 -0.008 -0.020 -0.007
## 13 SOCSUP1 =~ ss2t2 1.663 -0.081 -0.187 -0.096
## 14 SOCSUP1 =~ ss3t2 1.177 0.082 0.191 0.069
## 15 SOCSUP1 =~ se1t2 0.062 0.026 0.061 0.026
## 16 SOCSUP1 =~ se2t2 0.062 -0.022 -0.050 -0.022
## 17 SOCSUP1 =~ eb1t2 0.062 -0.055 -0.127 -0.025
## 18 SOCSUP1 =~ eb2t2 1.017 0.122 0.284 0.119
## 19 SOCSUP1 =~ eb3t2 1.886 -0.125 -0.290 -0.149
## 20 SELFEFF1 =~ ss1t1 0.008 0.012 0.015 0.006
## 21 SELFEFF1 =~ ss2t1 0.420 0.068 0.089 0.052
## 22 SELFEFF1 =~ ss3t1 0.467 -0.104 -0.137 -0.050
## 23 SELFEFF1 =~ eb1t1 1.336 0.494 0.647 0.155
## 24 SELFEFF1 =~ eb2t1 1.882 -0.328 -0.431 -0.205
## 25 SELFEFF1 =~ eb3t1 0.130 0.072 0.094 0.050
## 26 SELFEFF1 =~ ss1t2 0.014 0.017 0.023 0.009
## 27 SELFEFF1 =~ ss2t2 0.012 0.015 0.019 0.010
## 28 SELFEFF1 =~ ss3t2 0.041 -0.031 -0.041 -0.015
## 29 SELFEFF1 =~ se1t2 0.368 -0.171 -0.224 -0.096
## 30 SELFEFF1 =~ se2t2 0.368 0.141 0.184 0.083
## 31 SELFEFF1 =~ eb1t2 1.905 -0.666 -0.874 -0.175
## 32 SELFEFF1 =~ eb2t2 1.962 0.377 0.495 0.206
## 33 SELFEFF1 =~ eb3t2 0.001 0.006 0.008 0.004
## 34 EXBEH1 =~ ss1t1 1.608 -0.062 -0.184 -0.074
## 35 EXBEH1 =~ ss2t1 0.647 0.032 0.094 0.055
## 36 EXBEH1 =~ ss3t1 0.528 0.042 0.124 0.045
## 37 EXBEH1 =~ se1t1 1.275 -0.104 -0.306 -0.149
## 38 EXBEH1 =~ se2t1 1.275 0.085 0.251 0.121
## 39 EXBEH1 =~ ss1t2 0.000 0.001 0.003 0.001
## 40 EXBEH1 =~ ss2t2 0.005 -0.004 -0.011 -0.005
## 41 EXBEH1 =~ ss3t2 0.001 0.002 0.006 0.002
## 42 EXBEH1 =~ se1t2 0.013 0.014 0.041 0.017
## 43 EXBEH1 =~ se2t2 0.013 -0.011 -0.033 -0.015
## 44 EXBEH1 =~ eb1t2 7.801 -0.594 -1.754 -0.351
## 45 EXBEH1 =~ eb2t2 10.630 0.400 1.180 0.493
## 46 EXBEH1 =~ eb3t2 0.275 -0.047 -0.139 -0.071
## 47 SOCSUP2 =~ ss1t1 4.124 -0.128 -0.308 -0.124
## 48 SOCSUP2 =~ ss2t1 3.527 0.094 0.224 0.131
## 49 SOCSUP2 =~ ss3t1 0.342 0.043 0.102 0.037
## 50 SOCSUP2 =~ se1t1 0.008 0.010 0.023 0.011
## 51 SOCSUP2 =~ se2t1 0.008 -0.008 -0.019 -0.009
## 52 SOCSUP2 =~ eb1t1 6.082 0.461 1.106 0.265
## 53 SOCSUP2 =~ eb2t1 8.756 -0.312 -0.747 -0.356
## 54 SOCSUP2 =~ eb3t1 0.650 0.070 0.168 0.090
## 55 SOCSUP2 =~ se1t2 0.603 0.096 0.229 0.098
## 56 SOCSUP2 =~ se2t2 0.603 -0.078 -0.188 -0.084
## 57 SOCSUP2 =~ eb1t2 0.683 -0.187 -0.448 -0.090
## 58 SOCSUP2 =~ eb2t2 2.245 0.191 0.457 0.191
## 59 SOCSUP2 =~ eb3t2 1.381 -0.112 -0.269 -0.138
## 60 SELFEFF2 =~ ss1t1 3.519 -0.142 -0.271 -0.109
## 61 SELFEFF2 =~ ss2t1 2.110 0.088 0.168 0.098
## 62 SELFEFF2 =~ ss3t1 0.705 0.075 0.143 0.052
## 63 SELFEFF2 =~ se1t1 0.402 -0.099 -0.189 -0.092
## 64 SELFEFF2 =~ se2t1 0.402 0.082 0.156 0.075
## 65 SELFEFF2 =~ eb1t1 8.820 0.741 1.414 0.339
## 66 SELFEFF2 =~ eb2t1 12.105 -0.491 -0.936 -0.446
## 67 SELFEFF2 =~ eb3t1 0.618 0.092 0.176 0.094
## 68 SELFEFF2 =~ ss1t2 0.685 0.076 0.146 0.055
## 69 SELFEFF2 =~ ss2t2 0.433 -0.054 -0.103 -0.053
## 70 SELFEFF2 =~ ss3t2 0.139 -0.037 -0.070 -0.025
## 71 SELFEFF2 =~ eb1t2 6.132 -0.811 -1.547 -0.310
## 72 SELFEFF2 =~ eb2t2 7.608 0.514 0.980 0.409
## 73 SELFEFF2 =~ eb3t2 0.094 -0.043 -0.082 -0.042
## 74 EXBEH2 =~ ss1t1 3.668 -0.083 -0.268 -0.108
## 75 EXBEH2 =~ ss2t1 1.424 0.042 0.134 0.078
## 76 EXBEH2 =~ ss3t1 1.244 0.057 0.184 0.067
## 77 EXBEH2 =~ se1t1 1.176 -0.090 -0.291 -0.142
## 78 EXBEH2 =~ se2t1 1.176 0.074 0.239 0.115
## 79 EXBEH2 =~ eb1t1 11.295 0.530 1.702 0.408
## 80 EXBEH2 =~ eb2t1 15.441 -0.355 -1.139 -0.542
## 81 EXBEH2 =~ eb3t1 0.567 0.054 0.174 0.093
## 82 EXBEH2 =~ ss1t2 0.167 0.022 0.072 0.027
## 83 EXBEH2 =~ ss2t2 0.178 -0.020 -0.065 -0.033
## 84 EXBEH2 =~ ss3t2 0.012 -0.006 -0.021 -0.008
## 85 EXBEH2 =~ se1t2 0.173 0.053 0.170 0.073
## 86 EXBEH2 =~ se2t2 0.173 -0.044 -0.140 -0.063
## 87 ss1t1 ~~ ss2t1 1.250 0.233 0.233 0.310
## 88 ss1t1 ~~ ss3t1 0.182 0.150 0.150 0.132
## 89 ss1t1 ~~ se1t1 0.005 -0.015 -0.015 -0.011
## 90 ss1t1 ~~ se2t1 2.689 0.289 0.289 0.190
## 91 ss1t1 ~~ eb1t1 0.315 0.160 0.160 0.063
## 92 ss1t1 ~~ eb2t1 0.042 0.039 0.039 0.034
## 93 ss1t1 ~~ eb3t1 0.004 -0.012 -0.012 -0.008
## 94 ss1t1 ~~ ss2t2 1.889 -0.243 -0.243 -0.230
## 95 ss1t1 ~~ ss3t2 0.228 -0.112 -0.112 -0.106
## 96 ss1t1 ~~ se1t2 0.259 0.096 0.096 0.084
## 97 ss1t1 ~~ se2t2 2.552 -0.257 -0.257 -0.190
## 98 ss1t1 ~~ eb1t2 0.415 -0.227 -0.227 -0.069
## 99 ss1t1 ~~ eb2t2 0.063 -0.055 -0.055 -0.039
## 100 ss1t1 ~~ eb3t2 0.570 -0.153 -0.153 -0.098
## 101 ss2t1 ~~ ss3t1 2.147 -0.301 -0.301 -0.260
## 102 ss2t1 ~~ se1t1 1.504 0.206 0.206 0.149
## 103 ss2t1 ~~ se2t1 2.991 -0.251 -0.251 -0.161
## 104 ss2t1 ~~ eb1t1 0.000 -0.004 -0.004 -0.001
## 105 ss2t1 ~~ eb2t1 1.470 -0.193 -0.193 -0.162
## 106 ss2t1 ~~ eb3t1 3.361 0.301 0.301 0.197
## 107 ss2t1 ~~ ss1t2 0.163 -0.063 -0.063 -0.065
## 108 ss2t1 ~~ ss3t2 2.564 0.245 0.245 0.227
## 109 ss2t1 ~~ se1t2 0.903 0.148 0.148 0.126
## 110 ss2t1 ~~ se2t2 0.028 0.022 0.022 0.016
## 111 ss2t1 ~~ eb1t2 0.422 0.187 0.187 0.056
## 112 ss2t1 ~~ eb2t2 1.382 -0.212 -0.212 -0.147
## 113 ss2t1 ~~ eb3t2 0.072 -0.046 -0.046 -0.029
## 114 ss3t1 ~~ se1t1 0.476 -0.174 -0.174 -0.084
## 115 ss3t1 ~~ se2t1 0.557 -0.163 -0.163 -0.069
## 116 ss3t1 ~~ eb1t1 0.318 -0.197 -0.197 -0.051
## 117 ss3t1 ~~ eb2t1 0.080 0.067 0.067 0.038
## 118 ss3t1 ~~ eb3t1 0.181 -0.104 -0.104 -0.045
## 119 ss3t1 ~~ ss1t2 0.118 -0.093 -0.093 -0.064
## 120 ss3t1 ~~ ss2t2 0.943 0.194 0.194 0.119
## 121 ss3t1 ~~ se1t2 2.513 -0.371 -0.371 -0.209
## 122 ss3t1 ~~ se2t2 3.516 0.375 0.375 0.179
## 123 ss3t1 ~~ eb1t2 0.111 0.144 0.144 0.029
## 124 ss3t1 ~~ eb2t2 2.404 0.420 0.420 0.193
## 125 ss3t1 ~~ eb3t2 0.138 0.094 0.094 0.040
## sepc.nox
## 1 -0.046
## 2 -0.046
## 3 0.411
## 4 0.045
## 5 0.058
## 6 -0.374
## 7 0.070
## 8 -0.057
## 9 0.161
## 10 -0.265
## 11 0.161
## 12 -0.007
## 13 -0.096
## 14 0.069
## 15 0.026
## 16 -0.022
## 17 -0.025
## 18 0.119
## 19 -0.149
## 20 0.006
## 21 0.052
## 22 -0.050
## 23 0.155
## 24 -0.205
## 25 0.050
## 26 0.009
## 27 0.010
## 28 -0.015
## 29 -0.096
## 30 0.083
## 31 -0.175
## 32 0.206
## 33 0.004
## 34 -0.074
## 35 0.055
## 36 0.045
## 37 -0.149
## 38 0.121
## 39 0.001
## 40 -0.005
## 41 0.002
## 42 0.017
## 43 -0.015
## 44 -0.351
## 45 0.493
## 46 -0.071
## 47 -0.124
## 48 0.131
## 49 0.037
## 50 0.011
## 51 -0.009
## 52 0.265
## 53 -0.356
## 54 0.090
## 55 0.098
## 56 -0.084
## 57 -0.090
## 58 0.191
## 59 -0.138
## 60 -0.109
## 61 0.098
## 62 0.052
## 63 -0.092
## 64 0.075
## 65 0.339
## 66 -0.446
## 67 0.094
## 68 0.055
## 69 -0.053
## 70 -0.025
## 71 -0.310
## 72 0.409
## 73 -0.042
## 74 -0.108
## 75 0.078
## 76 0.067
## 77 -0.142
## 78 0.115
## 79 0.408
## 80 -0.542
## 81 0.093
## 82 0.027
## 83 -0.033
## 84 -0.008
## 85 0.073
## 86 -0.063
## 87 0.310
## 88 0.132
## 89 -0.011
## 90 0.190
## 91 0.063
## 92 0.034
## 93 -0.008
## 94 -0.230
## 95 -0.106
## 96 0.084
## 97 -0.190
## 98 -0.069
## 99 -0.039
## 100 -0.098
## 101 -0.260
## 102 0.149
## 103 -0.161
## 104 -0.001
## 105 -0.162
## 106 0.197
## 107 -0.065
## 108 0.227
## 109 0.126
## 110 0.016
## 111 0.056
## 112 -0.147
## 113 -0.029
## 114 -0.084
## 115 -0.069
## 116 -0.051
## 117 0.038
## 118 -0.045
## 119 -0.064
## 120 0.119
## 121 -0.209
## 122 0.179
## 123 0.029
## 124 0.193
## 125 0.040
## [ reached 'max' / getOption("max.print") -- omitted 71 rows ]DD.mod2 <- modindices(DD.fit2)with(DD.mod2, subset(DD.mod2[order(mi, decreasing = T), ], subset = mi >= 3.84))| lhs | op | rhs | mi | epc | sepc.lv | sepc.all | sepc.nox | |
|---|---|---|---|---|---|---|---|---|
| 140 | EXBEH2 | =~ | eb2t1 | 15.440864 | -0.3546176 | -1.1389684 | -0.5424548 | -0.5424548 |
| 126 | SELFEFF2 | =~ | eb2t1 | 12.104890 | -0.4908890 | -0.9364452 | -0.4459994 | -0.4459994 |
| 139 | EXBEH2 | =~ | eb1t1 | 11.295438 | 0.5298584 | 1.7018103 | 0.4078050 | 0.4078050 |
| 105 | EXBEH1 | =~ | eb2t2 | 10.629739 | 0.3998205 | 1.1804801 | 0.4927467 | 0.4927467 |
| 6 | EXBEH1 | =~ | eb1t1 | 8.986873 | 0.5808519 | 1.7149798 | 0.4109608 | 0.4109608 |
| 14 | EXBEH2 | =~ | eb1t2 | 8.986753 | -0.5808480 | -1.8655800 | -0.3738771 | -0.3738771 |
| 125 | SELFEFF2 | =~ | eb1t1 | 8.819876 | 0.7413010 | 1.4141440 | 0.3388715 | 0.3388715 |
| 113 | SOCSUP2 | =~ | eb2t1 | 8.755815 | -0.3119547 | -0.7473802 | -0.3559537 | -0.3559537 |
| 214 | eb1t1 | ~~ | eb2t2 | 7.869228 | 2.1474267 | 2.1474267 | 0.4436677 | 0.4436677 |
| 104 | EXBEH1 | =~ | eb1t2 | 7.801289 | -0.5939158 | -1.7535512 | -0.3514257 | -0.3514257 |
| 132 | SELFEFF2 | =~ | eb2t2 | 7.607759 | 0.5137326 | 0.9800229 | 0.4090734 | 0.4090734 |
| 195 | se1t1 | ~~ | eb2t2 | 7.131379 | -1.0178915 | -1.0178915 | -0.3942119 | -0.3942119 |
| 131 | SELFEFF2 | =~ | eb1t2 | 6.132218 | -0.8111754 | -1.5474400 | -0.3101193 | -0.3101193 |
| 112 | SOCSUP2 | =~ | eb1t1 | 6.082167 | 0.4614729 | 1.1055955 | 0.2649340 | 0.2649340 |
| 70 | SOCSUP1 | =~ | eb2t1 | 5.502834 | -0.2400151 | -0.5573404 | -0.2654437 | -0.2654437 |
| 229 | eb3t1 | ~~ | eb1t2 | 4.421558 | -1.1605709 | -1.1605709 | -0.1741365 | -0.1741365 |
| 199 | se2t1 | ~~ | eb3t1 | 4.403800 | -0.5683907 | -0.5683907 | -0.1832623 | -0.1832623 |
| 206 | se2t1 | ~~ | eb3t2 | 4.300079 | 0.5851247 | 0.5851247 | 0.1821014 | 0.1821014 |
| 107 | SOCSUP2 | =~ | ss1t1 | 4.124184 | -0.1284603 | -0.3077647 | -0.1243210 | -0.1243210 |
| 218 | eb2t1 | ~~ | ss2t2 | 3.930775 | 0.4275100 | 0.4275100 | 0.2561222 | 0.2561222 |
Fit a further modified measurement model
With the following the model syntax, we release the equality constraints imposed on the factor loading of item “eb2t1” and item “eb2t2”. To release the equality constraints, we simply remove the parameter labels.
DD.model3 <- '
SOCSUP1 =~ ss1t1 + a*ss2t1 + b*ss3t1
SELFEFF1 =~ se1t1 + c*se2t1
EXBEH1 =~ eb1t1 + eb2t1 + e*eb3t1
SOCSUP2 =~ ss1t2 + a*ss2t2 + b*ss3t2
SELFEFF2 =~ se1t2 + c*se2t2
EXBEH2 =~ eb1t2 + eb2t2 + e*eb3t2
ss1t1~~ss1t2
ss2t1~~ss2t2
ss3t1~~ss3t2
se1t1~~se1t2
se2t1~~se2t2
eb1t1~~eb1t2
eb2t1~~eb2t2
eb3t1~~eb3t2
'
DD.fit3 <- sem(DD.model3, sample.cov = covmat, sample.nobs = 84)
summary(DD.fit3, fit.measures = T, standardized = T)## lavaan 0.6-19 ended normally after 167 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 55
## Number of equality constraints 4
##
## Number of observations 84
##
## Model Test User Model:
##
## Test statistic 91.571
## Degrees of freedom 85
## P-value (Chi-square) 0.294
##
## Model Test Baseline Model:
##
## Test statistic 904.195
## Degrees of freedom 120
## P-value 0.000
##
## User Model versus Baseline Model:
##
## Comparative Fit Index (CFI) 0.992
## Tucker-Lewis Index (TLI) 0.988
##
## Loglikelihood and Information Criteria:
##
## Loglikelihood user model (H0) -2687.524
## Loglikelihood unrestricted model (H1) -2641.739
##
## Akaike (AIC) 5477.048
## Bayesian (BIC) 5601.020
## Sample-size adjusted Bayesian (SABIC) 5440.139
##
## Root Mean Square Error of Approximation:
##
## RMSEA 0.030
## 90 Percent confidence interval - lower 0.000
## 90 Percent confidence interval - upper 0.069
## P-value H_0: RMSEA <= 0.050 0.759
## P-value H_0: RMSEA >= 0.080 0.012
##
## Standardized Root Mean Square Residual:
##
## SRMR 0.063
##
## Parameter Estimates:
##
## Standard errors Standard
## Information Expected
## Information saturated (h1) model Structured
##
## Latent Variables:
## Estimate Std.Err z-value P(>|z|)
## SOCSUP1 =~
## ss1t1 1.000
## ss2t1 (a) 0.632 0.048 13.290 0.000
## ss3t1 (b) 1.026 0.070 14.634 0.000
## SELFEFF1 =~
## se1t1 1.000
## se2t1 (c) 0.768 0.133 5.773 0.000
## EXBEH1 =~
## eb1t1 1.000
## eb2t1 0.349 0.075 4.643 0.000
## eb3t1 (e) 0.225 0.060 3.735 0.000
## SOCSUP2 =~
## ss1t2 1.000
## ss2t2 (a) 0.632 0.048 13.290 0.000
## ss3t2 (b) 1.026 0.070 14.634 0.000
## SELFEFF2 =~
## se1t2 1.000
## se2t2 (c) 0.768 0.133 5.773 0.000
## EXBEH2 =~
## eb1t2 1.000
## eb2t2 0.775 0.143 5.431 0.000
## eb3t2 (e) 0.225 0.060 3.735 0.000
## Std.lv Std.all
##
## 2.324 0.939
## 1.468 0.858
## 2.385 0.874
##
## 1.357 0.669
## 1.043 0.497
##
## 3.339 0.783
## 1.165 0.617
## 0.751 0.396
##
## 2.397 0.910
## 1.514 0.775
## 2.460 0.893
##
## 1.963 0.839
## 1.508 0.682
##
## 2.959 0.617
## 2.292 0.867
## 0.665 0.346
##
## Covariances:
## Estimate Std.Err z-value P(>|z|)
## .ss1t1 ~~
## .ss1t2 0.427 0.226 1.891 0.059
## .ss2t1 ~~
## .ss2t2 0.396 0.153 2.589 0.010
## .ss3t1 ~~
## .ss3t2 0.715 0.290 2.469 0.014
## .se1t1 ~~
## .se1t2 0.532 0.467 1.140 0.254
## .se2t1 ~~
## .se2t2 2.000 0.457 4.372 0.000
## .eb1t1 ~~
## .eb1t2 7.232 1.805 4.006 0.000
## .eb2t1 ~~
## .eb2t2 0.027 0.381 0.072 0.943
## .eb3t1 ~~
## .eb3t2 1.052 0.374 2.815 0.005
## SOCSUP1 ~~
## SELFEFF1 1.448 0.511 2.834 0.005
## EXBEH1 3.678 1.113 3.305 0.001
## SOCSUP2 4.162 0.855 4.869 0.000
## SELFEFF2 1.970 0.634 3.109 0.002
## EXBEH2 2.998 1.042 2.877 0.004
## SELFEFF1 ~~
## EXBEH1 2.429 0.822 2.956 0.003
## SOCSUP2 1.227 0.522 2.351 0.019
## SELFEFF2 1.666 0.600 2.777 0.005
## EXBEH2 1.709 0.739 2.311 0.021
## EXBEH1 ~~
## SOCSUP2 4.857 1.241 3.915 0.000
## SELFEFF2 4.218 1.119 3.769 0.000
## EXBEH2 8.839 2.625 3.367 0.001
## SOCSUP2 ~~
## SELFEFF2 2.608 0.692 3.768 0.000
## EXBEH2 4.437 1.241 3.576 0.000
## SELFEFF2 ~~
## EXBEH2 4.564 1.198 3.809 0.000
## Std.lv Std.all
##
## 0.427 0.458
##
## 0.396 0.366
##
## 0.715 0.434
##
## 0.532 0.277
##
## 2.000 0.680
##
## 7.232 0.723
##
## 0.027 0.014
##
## 1.052 0.334
##
## 0.459 0.459
## 0.474 0.474
## 0.747 0.747
## 0.432 0.432
## 0.436 0.436
##
## 0.536 0.536
## 0.377 0.377
## 0.625 0.625
## 0.425 0.425
##
## 0.607 0.607
## 0.643 0.643
## 0.895 0.895
##
## 0.554 0.554
## 0.626 0.626
##
## 0.785 0.785
##
## Variances:
## Estimate Std.Err z-value P(>|z|)
## .ss1t1 0.725 0.263 2.762 0.006
## .ss2t1 0.769 0.154 4.983 0.000
## .ss3t1 1.762 0.374 4.705 0.000
## .se1t1 2.272 0.663 3.424 0.001
## .se2t1 3.305 0.606 5.457 0.000
## .eb1t1 7.035 1.963 3.584 0.000
## .eb2t1 2.211 0.428 5.167 0.000
## .eb3t1 3.034 0.486 6.239 0.000
## .ss1t2 1.196 0.345 3.464 0.001
## .ss2t2 1.526 0.272 5.601 0.000
## .ss3t2 1.543 0.391 3.944 0.000
## .se1t2 1.623 0.607 2.673 0.008
## .se2t2 2.619 0.523 5.009 0.000
## .eb1t2 14.218 2.497 5.695 0.000
## .eb2t2 1.739 0.737 2.361 0.018
## .eb3t2 3.265 0.515 6.341 0.000
## SOCSUP1 5.399 0.952 5.672 0.000
## SELFEFF1 1.842 0.666 2.766 0.006
## EXBEH1 11.147 2.982 3.739 0.000
## SOCSUP2 5.744 1.046 5.493 0.000
## SELFEFF2 3.855 0.968 3.980 0.000
## EXBEH2 8.758 2.961 2.958 0.003
## Std.lv Std.all
## 0.725 0.118
## 0.769 0.263
## 1.762 0.237
## 2.272 0.552
## 3.305 0.753
## 7.035 0.387
## 2.211 0.620
## 3.034 0.843
## 1.196 0.172
## 1.526 0.399
## 1.543 0.203
## 1.623 0.296
## 2.619 0.535
## 14.218 0.619
## 1.739 0.249
## 3.265 0.881
## 1.000 1.000
## 1.000 1.000
## 1.000 1.000
## 1.000 1.000
## 1.000 1.000
## 1.000 1.000DD.mod3 <- modindices(DD.fit3)## Warning: lavaan->modindices():
## the modindices() function ignores equality
## constraints; use lavTestScore() to assess the impact
## of releasing one or multiple constraints.with(DD.mod3, subset(DD.mod3[order(mi, decreasing = T), ], subset = mi >= 3.84))## lhs op rhs mi epc sepc.lv sepc.all
## 194 se1t1 ~~ eb2t2 6.073 -0.979 -0.979 -0.493
## 205 se2t1 ~~ eb3t2 4.283 0.579 0.579 0.176
## 197 se2t1 ~~ eb2t1 4.131 0.527 0.527 0.195
## 106 SOCSUP2 =~ ss1t1 4.004 -0.127 -0.304 -0.123
## 213 eb1t1 ~~ eb2t2 3.932 2.004 2.004 0.573
## 198 se2t1 ~~ eb3t1 3.926 -0.538 -0.538 -0.170
## sepc.nox
## 194 -0.493
## 205 0.176
## 197 0.195
## 106 -0.123
## 213 0.573
## 198 -0.170The final measurement model
semPaths(DD.fit3, title = FALSE, curvePivot = TRUE, intercepts = F, optimizeLatRes = T)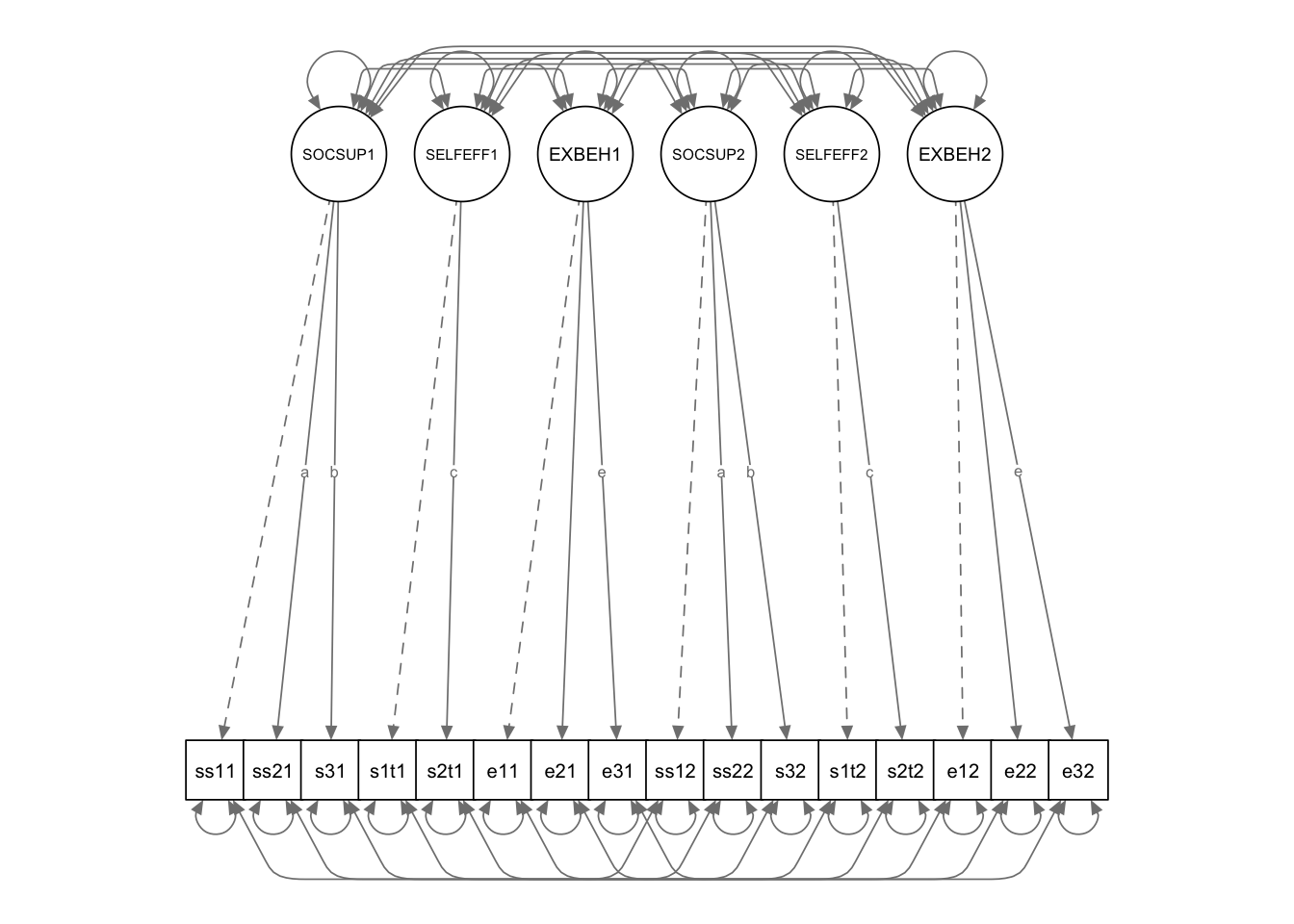
Step 2: Structural phase
Fit the initial structural model
DD.model4 <- '
# measurement model
SOCSUP1 =~ ss1t1 + a*ss2t1 + b*ss3t1
SELFEFF1 =~ se1t1 + c*se2t1
EXBEH1 =~ eb1t1 + eb2t1 + e*eb3t1
SOCSUP2 =~ ss1t2 + a*ss2t2 + b*ss3t2
SELFEFF2 =~ se1t2 + c*se2t2
EXBEH2 =~ eb1t2 + eb2t2 + e*eb3t2
ss1t1~~ss1t2
ss2t1~~ss2t2
ss3t1~~ss3t2
se1t1~~se1t2
se2t1~~se2t2
eb1t1~~eb1t2
eb2t1~~eb2t2
eb3t1~~eb3t2
# structural model
EXBEH2 ~ EXBEH1 + SELFEFF1 + SELFEFF2
SELFEFF2 ~ SELFEFF1 + SOCSUP1 + SOCSUP2
SOCSUP2 ~ SOCSUP1
'
DD.fit4 <- sem(DD.model4, sample.cov = covmat, sample.nobs = 84)
summary(DD.fit4, fit.measures = T, standardized = T)## lavaan 0.6-19 ended normally after 122 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 50
## Number of equality constraints 4
##
## Number of observations 84
##
## Model Test User Model:
##
## Test statistic 103.825
## Degrees of freedom 90
## P-value (Chi-square) 0.151
##
## Model Test Baseline Model:
##
## Test statistic 904.195
## Degrees of freedom 120
## P-value 0.000
##
## User Model versus Baseline Model:
##
## Comparative Fit Index (CFI) 0.982
## Tucker-Lewis Index (TLI) 0.976
##
## Loglikelihood and Information Criteria:
##
## Loglikelihood user model (H0) -2693.651
## Loglikelihood unrestricted model (H1) -2641.739
##
## Akaike (AIC) 5479.302
## Bayesian (BIC) 5591.120
## Sample-size adjusted Bayesian (SABIC) 5446.012
##
## Root Mean Square Error of Approximation:
##
## RMSEA 0.043
## 90 Percent confidence interval - lower 0.000
## 90 Percent confidence interval - upper 0.076
## P-value H_0: RMSEA <= 0.050 0.608
## P-value H_0: RMSEA >= 0.080 0.028
##
## Standardized Root Mean Square Residual:
##
## SRMR 0.090
##
## Parameter Estimates:
##
## Standard errors Standard
## Information Expected
## Information saturated (h1) model Structured
##
## Latent Variables:
## Estimate Std.Err z-value P(>|z|)
## SOCSUP1 =~
## ss1t1 1.000
## ss2t1 (a) 0.632 0.048 13.263 0.000
## ss3t1 (b) 1.027 0.071 14.513 0.000
## SELFEFF1 =~
## se1t1 1.000
## se2t1 (c) 0.783 0.139 5.624 0.000
## EXBEH1 =~
## eb1t1 1.000
## eb2t1 0.415 0.094 4.411 0.000
## eb3t1 (e) 0.255 0.068 3.724 0.000
## SOCSUP2 =~
## ss1t2 1.000
## ss2t2 (a) 0.632 0.048 13.263 0.000
## ss3t2 (b) 1.027 0.071 14.513 0.000
## SELFEFF2 =~
## se1t2 1.000
## se2t2 (c) 0.783 0.139 5.624 0.000
## EXBEH2 =~
## eb1t2 1.000
## eb2t2 0.942 0.211 4.452 0.000
## eb3t2 (e) 0.255 0.068 3.724 0.000
## Std.lv Std.all
##
## 2.320 0.935
## 1.468 0.859
## 2.382 0.875
##
## 1.252 0.614
## 0.980 0.468
##
## 3.032 0.718
## 1.259 0.667
## 0.772 0.405
##
## 2.407 0.913
## 1.522 0.776
## 2.470 0.892
##
## 1.951 0.844
## 1.528 0.685
##
## 2.468 0.533
## 2.324 0.914
## 0.628 0.330
##
## Regressions:
## Estimate Std.Err z-value P(>|z|)
## EXBEH2 ~
## EXBEH1 0.712 0.191 3.736 0.000
## SELFEFF1 -1.017 0.562 -1.810 0.070
## SELFEFF2 0.802 0.253 3.172 0.002
## SELFEFF2 ~
## SELFEFF1 1.015 0.278 3.646 0.000
## SOCSUP1 -0.189 0.143 -1.323 0.186
## SOCSUP2 0.367 0.125 2.931 0.003
## SOCSUP2 ~
## SOCSUP1 0.782 0.085 9.208 0.000
## Std.lv Std.all
##
## 0.875 0.875
## -0.516 -0.516
## 0.634 0.634
##
## 0.651 0.651
## -0.225 -0.225
## 0.452 0.452
##
## 0.754 0.754
##
## Covariances:
## Estimate Std.Err z-value P(>|z|)
## .ss1t1 ~~
## .ss1t2 0.422 0.229 1.844 0.065
## .ss2t1 ~~
## .ss2t2 0.386 0.153 2.517 0.012
## .ss3t1 ~~
## .ss3t2 0.716 0.292 2.448 0.014
## .se1t1 ~~
## .se1t2 0.493 0.471 1.046 0.295
## .se2t1 ~~
## .se2t2 2.010 0.466 4.316 0.000
## .eb1t1 ~~
## .eb1t2 8.531 1.932 4.416 0.000
## .eb2t1 ~~
## .eb2t2 -0.255 0.437 -0.584 0.559
## .eb3t1 ~~
## .eb3t2 1.042 0.371 2.806 0.005
## SOCSUP1 ~~
## SELFEFF1 1.442 0.484 2.978 0.003
## EXBEH1 3.307 1.040 3.180 0.001
## SELFEFF1 ~~
## EXBEH1 2.459 0.766 3.208 0.001
## Std.lv Std.all
##
## 0.422 0.449
##
## 0.386 0.357
##
## 0.716 0.435
##
## 0.493 0.247
##
## 2.010 0.668
##
## 8.531 0.742
##
## -0.255 -0.176
##
## 1.042 0.333
##
## 0.497 0.497
## 0.470 0.470
##
## 0.648 0.648
##
## Variances:
## Estimate Std.Err z-value P(>|z|)
## .ss1t1 0.769 0.265 2.908 0.004
## .ss2t1 0.763 0.154 4.965 0.000
## .ss3t1 1.735 0.371 4.673 0.000
## .se1t1 2.587 0.631 4.102 0.000
## .se2t1 3.427 0.605 5.669 0.000
## .eb1t1 8.619 2.042 4.221 0.000
## .eb2t1 1.977 0.459 4.305 0.000
## .eb3t1 3.026 0.487 6.220 0.000
## .ss1t2 1.149 0.351 3.270 0.001
## .ss2t2 1.535 0.275 5.586 0.000
## .ss3t2 1.563 0.403 3.877 0.000
## .se1t2 1.543 0.620 2.489 0.013
## .se2t2 2.641 0.542 4.874 0.000
## .eb1t2 15.353 2.589 5.929 0.000
## .eb2t2 1.061 0.934 1.137 0.256
## .eb3t2 3.235 0.508 6.364 0.000
## SOCSUP1 5.384 0.954 5.643 0.000
## SELFEFF1 1.567 0.599 2.614 0.009
## EXBEH1 9.193 2.703 3.401 0.001
## .SOCSUP2 2.503 0.495 5.059 0.000
## .SELFEFF2 1.520 0.545 2.792 0.005
## .EXBEH2 0.527 0.786 0.671 0.502
## Std.lv Std.all
## 0.769 0.125
## 0.763 0.262
## 1.735 0.234
## 2.587 0.623
## 3.427 0.781
## 8.619 0.484
## 1.977 0.555
## 3.026 0.836
## 1.149 0.166
## 1.535 0.399
## 1.563 0.204
## 1.543 0.288
## 2.641 0.531
## 15.353 0.716
## 1.061 0.164
## 3.235 0.891
## 1.000 1.000
## 1.000 1.000
## 1.000 1.000
## 0.432 0.432
## 0.399 0.399
## 0.087 0.087Examine momdel fit and modification indicies
The model fit looks acceptable. We do not have enough evidence to reject the model.
DD.mod4 <- modindices(DD.fit4)## Warning: lavaan->modindices():
## the modindices() function ignores equality
## constraints; use lavTestScore() to assess the impact
## of releasing one or multiple constraints.with(DD.mod4, subset(DD.mod4[order(mi, decreasing = T), ], subset = mi >= 3.84))## lhs op rhs mi epc sepc.lv sepc.all
## 259 EXBEH1 ~~ SOCSUP2 8.817 1.601 0.334 0.334
## 275 EXBEH1 ~ SOCSUP2 8.817 0.640 0.508 0.508
## 271 SOCSUP2 ~ EXBEH1 8.427 0.238 0.300 0.300
## 269 SOCSUP2 ~ EXBEH2 8.404 0.340 0.349 0.349
## 274 EXBEH1 ~ SELFEFF2 7.606 1.038 0.668 0.668
## 273 EXBEH1 ~ EXBEH2 7.583 1.288 1.048 1.048
## 284 SOCSUP1 ~ SELFEFF2 6.330 -2.075 -1.744 -1.744
## 133 EXBEH2 =~ eb1t1 6.021 0.720 1.778 0.421
## 128 EXBEH2 =~ ss1t1 5.396 -0.130 -0.321 -0.129
## 253 SOCSUP1 ~~ SOCSUP2 5.381 -2.001 -0.545 -0.545
## 285 SOCSUP1 ~ SOCSUP2 5.381 -0.800 -0.829 -0.829
## 193 se2t1 ~~ eb3t1 5.344 -0.632 -0.632 -0.196
## 189 se1t1 ~~ eb2t2 5.261 -0.914 -0.914 -0.552
## 101 SOCSUP2 =~ ss1t1 5.021 -0.143 -0.345 -0.139
## 86 SELFEFF1 =~ eb2t2 4.442 -1.184 -1.482 -0.583
## 114 SELFEFF2 =~ ss1t1 4.345 -0.155 -0.302 -0.122
## 222 eb3t1 ~~ se2t2 4.008 0.499 0.499 0.176
## 200 se2t1 ~~ eb3t2 3.988 0.556 0.556 0.167
## 192 se2t1 ~~ eb2t1 3.972 0.520 0.520 0.200
## 208 eb1t1 ~~ eb2t2 3.854 2.041 2.041 0.675
## sepc.nox
## 259 0.334
## 275 0.508
## 271 0.300
## 269 0.349
## 274 0.668
## 273 1.048
## 284 -1.744
## 133 0.421
## 128 -0.129
## 253 -0.545
## 285 -0.829
## 193 -0.196
## 189 -0.552
## 101 -0.139
## 86 -0.583
## 114 -0.122
## 222 0.176
## 200 0.167
## 192 0.200
## 208 0.675Final structural model
semPaths(DD.fit4, title = FALSE, curvePivot = TRUE, intercepts = F, optimizeLatRes = T)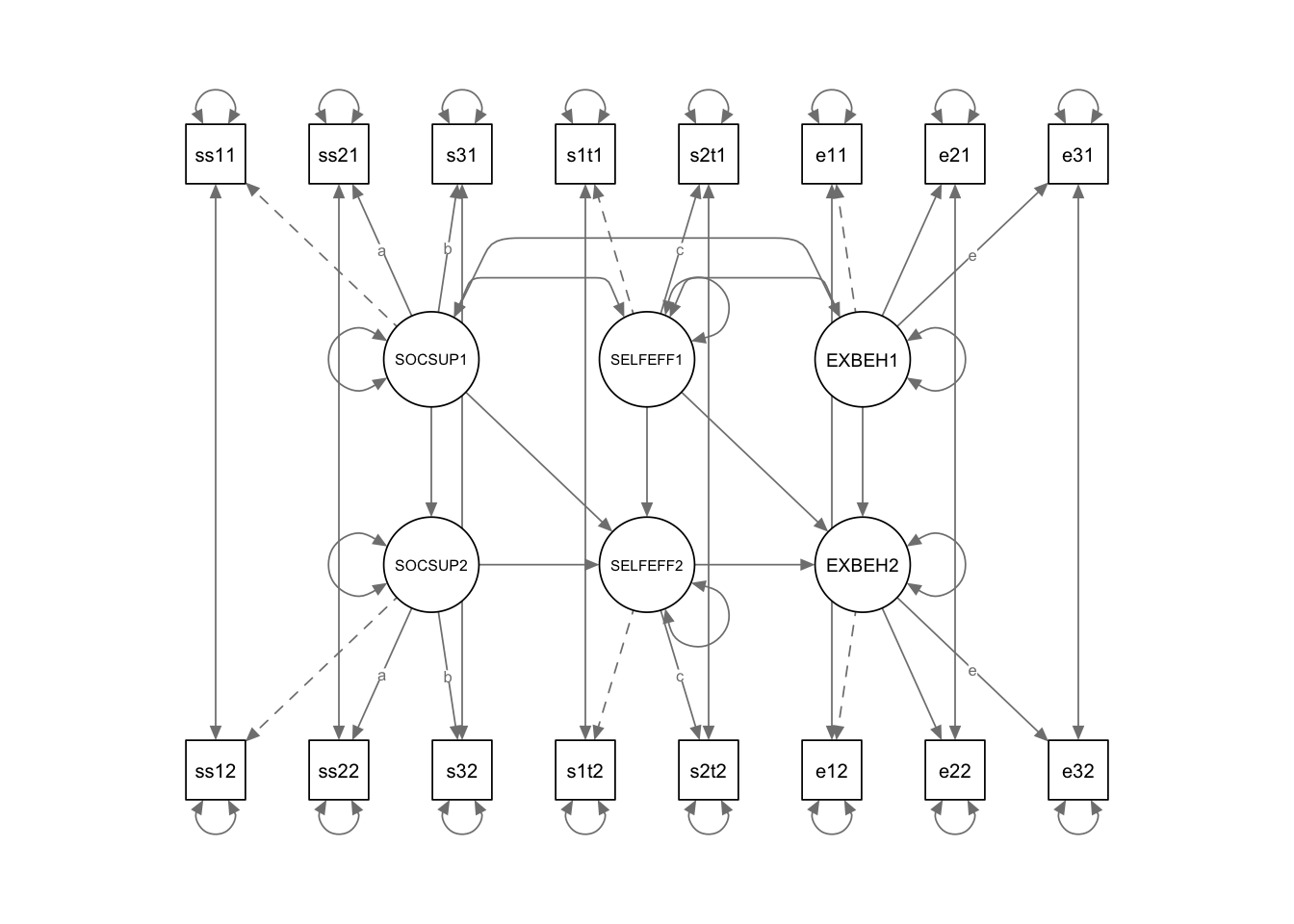
© Copyright 2025 @Yi Feng and @Gregory R. Hancock.