Nonlinear Latent Growth Models
In this tutorial, we are going to use lavaan to fit
nonlinear latent growth models.
Load the pacakges
library(lavaan)
library(semPlot)Example: Air Data
Read the data
lower <- '
93.650
79.637 93.564
72.890 87.718 95.732
62.623 76.941 81.886 86.054
53.342 63.803 69.612 70.611 73.666
43.820 50.644 54.443 56.446 58.353 59.154
35.183 41.099 46.016 45.494 49.900 46.163 54.169
39.839 44.099 46.105 44.074 48.345 45.847 49.329 60.528
35.497 39.169 42.262 39.429 42.954 42.950 47.120 54.570 66.183
'
smeans <- c(20.121, 25.521, 29.321, 32.400, 34.186, 35.600, 37.729, 39.029, 38.786)
covmat <- getCov(lower)
rownames(covmat) <- colnames(covmat) <- paste0('V', 1:9)Linear Model
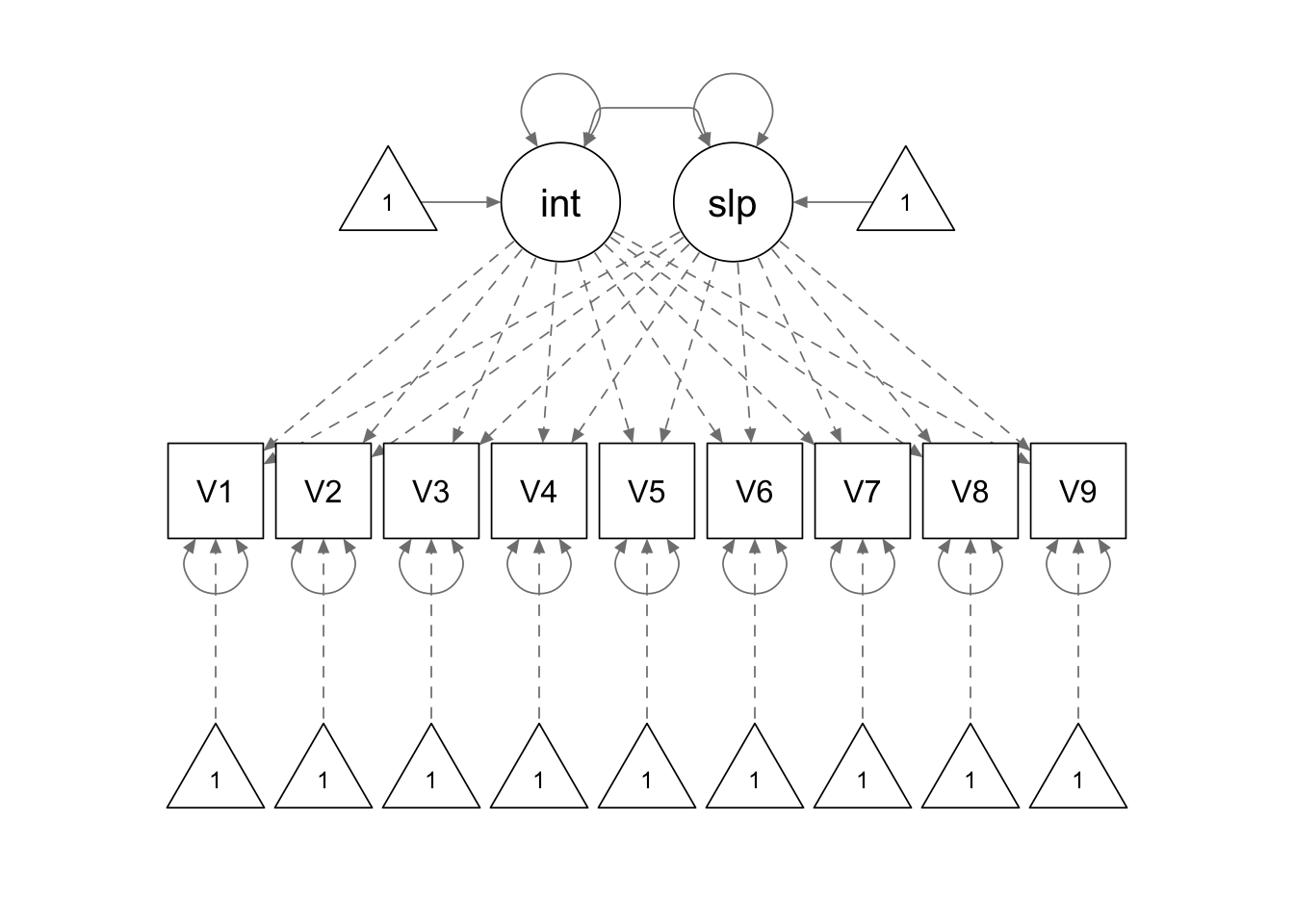
linear.model <- '
interc =~ 1*V1 + 1*V2 + 1*V3 + 1*V4 + 1*V5 + 1*V6 + 1*V7 + 1*V8 + 1*V9
slope =~ 0*V1 + 1*V2 + 2*V3 + 3*V4 + 4*V5 + 5*V6 + 6*V7 + 7*V8 + 8*V9
'
linear.fit <- growth(linear.model, sample.cov=covmat, sample.mean=smeans, sample.nobs = 140)
summary(linear.fit, fit.measures = T)## lavaan 0.6.15 ended normally after 91 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 14
##
## Number of observations 140
##
## Model Test User Model:
##
## Test statistic 413.528
## Degrees of freedom 40
## P-value (Chi-square) 0.000
##
## Model Test Baseline Model:
##
## Test statistic 1702.962
## Degrees of freedom 36
## P-value 0.000
##
## User Model versus Baseline Model:
##
## Comparative Fit Index (CFI) 0.776
## Tucker-Lewis Index (TLI) 0.798
##
## Loglikelihood and Information Criteria:
##
## Loglikelihood user model (H0) -3851.973
## Loglikelihood unrestricted model (H1) -3645.210
##
## Akaike (AIC) 7731.947
## Bayesian (BIC) 7773.130
## Sample-size adjusted Bayesian (SABIC) 7728.836
##
## Root Mean Square Error of Approximation:
##
## RMSEA 0.258
## 90 Percent confidence interval - lower 0.236
## 90 Percent confidence interval - upper 0.281
## P-value H_0: RMSEA <= 0.050 0.000
## P-value H_0: RMSEA >= 0.080 1.000
##
## Standardized Root Mean Square Residual:
##
## SRMR 0.165
##
## Parameter Estimates:
##
## Standard errors Standard
## Information Expected
## Information saturated (h1) model Structured
##
## Latent Variables:
## Estimate Std.Err z-value P(>|z|)
## interc =~
## V1 1.000
## V2 1.000
## V3 1.000
## V4 1.000
## V5 1.000
## V6 1.000
## V7 1.000
## V8 1.000
## V9 1.000
## slope =~
## V1 0.000
## V2 1.000
## V3 2.000
## V4 3.000
## V5 4.000
## V6 5.000
## V7 6.000
## V8 7.000
## V9 8.000
##
## Covariances:
## Estimate Std.Err z-value P(>|z|)
## interc ~~
## slope -9.629 1.526 -6.311 0.000
##
## Intercepts:
## Estimate Std.Err z-value P(>|z|)
## .V1 0.000
## .V2 0.000
## .V3 0.000
## .V4 0.000
## .V5 0.000
## .V6 0.000
## .V7 0.000
## .V8 0.000
## .V9 0.000
## interc 24.599 0.902 27.279 0.000
## slope 2.114 0.115 18.443 0.000
##
## Variances:
## Estimate Std.Err z-value P(>|z|)
## .V1 59.014 7.787 7.579 0.000
## .V2 11.063 2.057 5.378 0.000
## .V3 8.568 1.486 5.764 0.000
## .V4 14.757 2.002 7.372 0.000
## .V5 12.952 1.733 7.473 0.000
## .V6 12.023 1.639 7.334 0.000
## .V7 8.774 1.375 6.379 0.000
## .V8 8.675 1.612 5.381 0.000
## .V9 25.644 3.728 6.878 0.000
## interc 107.692 13.615 7.910 0.000
## slope 1.574 0.220 7.139 0.000Quadratic Model
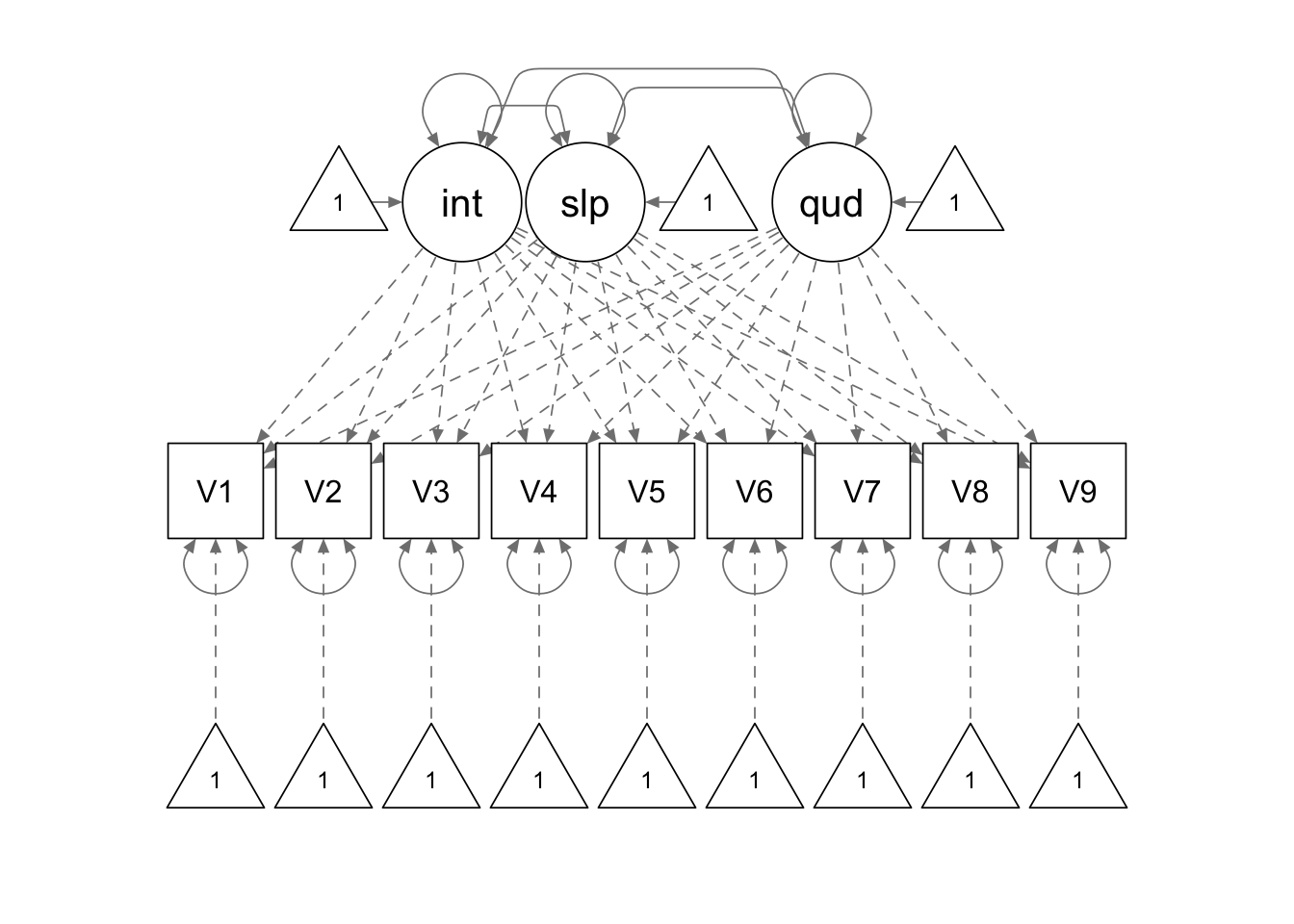
quad.model <- '
interc =~ 1*V1 + 1*V2 + 1*V3 + 1*V4 + 1*V5 + 1*V6 + 1*V7 + 1*V8 + 1*V9
slope =~ 0*V1 + 1*V2 + 2*V3 + 3*V4 + 4*V5 + 5*V6 + 6*V7 + 7*V8 + 8*V9
quad =~ 0*V1 + 1*V2 + 4*V3 + 9*V4 + 16*V5 + 25*V6 + 36*V7 + 49*V8 + 64*V9
'
quad.fit <- growth(quad.model, sample.cov=covmat, sample.mean=smeans, sample.nobs = 140)
summary(quad.fit, fit.measures = T)## lavaan 0.6.15 ended normally after 108 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 18
##
## Number of observations 140
##
## Model Test User Model:
##
## Test statistic 154.710
## Degrees of freedom 36
## P-value (Chi-square) 0.000
##
## Model Test Baseline Model:
##
## Test statistic 1702.962
## Degrees of freedom 36
## P-value 0.000
##
## User Model versus Baseline Model:
##
## Comparative Fit Index (CFI) 0.929
## Tucker-Lewis Index (TLI) 0.929
##
## Loglikelihood and Information Criteria:
##
## Loglikelihood user model (H0) -3722.565
## Loglikelihood unrestricted model (H1) -3645.210
##
## Akaike (AIC) 7481.129
## Bayesian (BIC) 7534.079
## Sample-size adjusted Bayesian (SABIC) 7477.129
##
## Root Mean Square Error of Approximation:
##
## RMSEA 0.153
## 90 Percent confidence interval - lower 0.129
## 90 Percent confidence interval - upper 0.179
## P-value H_0: RMSEA <= 0.050 0.000
## P-value H_0: RMSEA >= 0.080 1.000
##
## Standardized Root Mean Square Residual:
##
## SRMR 0.104
##
## Parameter Estimates:
##
## Standard errors Standard
## Information Expected
## Information saturated (h1) model Structured
##
## Latent Variables:
## Estimate Std.Err z-value P(>|z|)
## interc =~
## V1 1.000
## V2 1.000
## V3 1.000
## V4 1.000
## V5 1.000
## V6 1.000
## V7 1.000
## V8 1.000
## V9 1.000
## slope =~
## V1 0.000
## V2 1.000
## V3 2.000
## V4 3.000
## V5 4.000
## V6 5.000
## V7 6.000
## V8 7.000
## V9 8.000
## quad =~
## V1 0.000
## V2 1.000
## V3 4.000
## V4 9.000
## V5 16.000
## V6 25.000
## V7 36.000
## V8 49.000
## V9 64.000
##
## Covariances:
## Estimate Std.Err z-value P(>|z|)
## interc ~~
## slope -15.556 3.359 -4.631 0.000
## quad 0.777 0.336 2.315 0.021
## slope ~~
## quad -0.730 0.129 -5.674 0.000
##
## Intercepts:
## Estimate Std.Err z-value P(>|z|)
## .V1 0.000
## .V2 0.000
## .V3 0.000
## .V4 0.000
## .V5 0.000
## .V6 0.000
## .V7 0.000
## .V8 0.000
## .V9 0.000
## interc 21.348 0.918 23.260 0.000
## slope 4.347 0.267 16.257 0.000
## quad -0.269 0.029 -9.188 0.000
##
## Variances:
## Estimate Std.Err z-value P(>|z|)
## .V1 29.757 4.655 6.393 0.000
## .V2 3.418 1.264 2.705 0.007
## .V3 9.360 1.350 6.935 0.000
## .V4 10.315 1.472 7.008 0.000
## .V5 6.778 1.140 5.946 0.000
## .V6 10.089 1.485 6.794 0.000
## .V7 9.474 1.377 6.882 0.000
## .V8 8.254 1.416 5.828 0.000
## .V9 7.344 2.441 3.009 0.003
## interc 111.181 14.160 7.852 0.000
## slope 7.587 1.220 6.221 0.000
## quad 0.088 0.015 5.902 0.000Spline Model-1
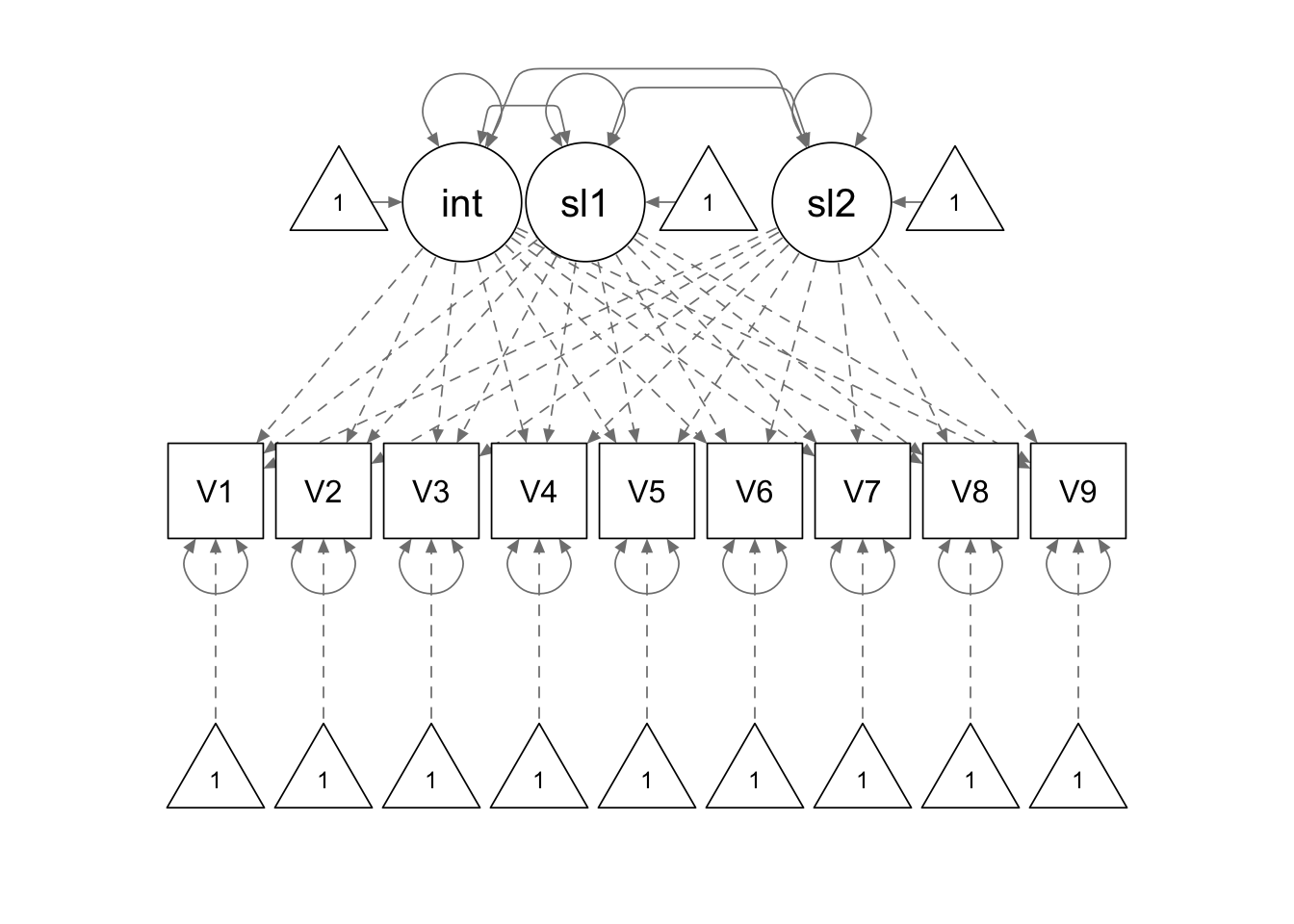
spline.model1 <- '
interc =~ 1*V1 + 1*V2 + 1*V3 + 1*V4 + 1*V5 + 1*V6 + 1*V7 + 1*V8 + 1*V9
slope1 =~ 0*V1 + 1*V2 + 2*V3 + 3*V4 + 4*V5 + 4*V6 + 4*V7 + 4*V8 + 4*V9
slope2 =~ 0*V1 + 0*V2 + 0*V3 + 0*V4 + 0*V5 + 1*V6 + 2*V7 + 3*V8 + 4*V9
'
spline.fit1 <- growth(spline.model1, sample.cov = covmat, sample.mean = smeans, sample.nobs = 140)
summary(spline.fit1, fit.measures = T)## lavaan 0.6.15 ended normally after 117 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 18
##
## Number of observations 140
##
## Model Test User Model:
##
## Test statistic 172.011
## Degrees of freedom 36
## P-value (Chi-square) 0.000
##
## Model Test Baseline Model:
##
## Test statistic 1702.962
## Degrees of freedom 36
## P-value 0.000
##
## User Model versus Baseline Model:
##
## Comparative Fit Index (CFI) 0.918
## Tucker-Lewis Index (TLI) 0.918
##
## Loglikelihood and Information Criteria:
##
## Loglikelihood user model (H0) -3731.215
## Loglikelihood unrestricted model (H1) -3645.210
##
## Akaike (AIC) 7498.430
## Bayesian (BIC) 7551.379
## Sample-size adjusted Bayesian (SABIC) 7494.430
##
## Root Mean Square Error of Approximation:
##
## RMSEA 0.164
## 90 Percent confidence interval - lower 0.140
## 90 Percent confidence interval - upper 0.189
## P-value H_0: RMSEA <= 0.050 0.000
## P-value H_0: RMSEA >= 0.080 1.000
##
## Standardized Root Mean Square Residual:
##
## SRMR 0.119
##
## Parameter Estimates:
##
## Standard errors Standard
## Information Expected
## Information saturated (h1) model Structured
##
## Latent Variables:
## Estimate Std.Err z-value P(>|z|)
## interc =~
## V1 1.000
## V2 1.000
## V3 1.000
## V4 1.000
## V5 1.000
## V6 1.000
## V7 1.000
## V8 1.000
## V9 1.000
## slope1 =~
## V1 0.000
## V2 1.000
## V3 2.000
## V4 3.000
## V5 4.000
## V6 4.000
## V7 4.000
## V8 4.000
## V9 4.000
## slope2 =~
## V1 0.000
## V2 0.000
## V3 0.000
## V4 0.000
## V5 0.000
## V6 1.000
## V7 2.000
## V8 3.000
## V9 4.000
##
## Covariances:
## Estimate Std.Err z-value P(>|z|)
## interc ~~
## slope1 -12.985 2.243 -5.790 0.000
## slope2 -6.891 1.807 -3.813 0.000
## slope1 ~~
## slope2 -0.125 0.335 -0.374 0.708
##
## Intercepts:
## Estimate Std.Err z-value P(>|z|)
## .V1 0.000
## .V2 0.000
## .V3 0.000
## .V4 0.000
## .V5 0.000
## .V6 0.000
## .V7 0.000
## .V8 0.000
## .V9 0.000
## interc 22.458 0.908 24.727 0.000
## slope1 3.048 0.172 17.693 0.000
## slope2 1.269 0.160 7.937 0.000
##
## Variances:
## Estimate Std.Err z-value P(>|z|)
## .V1 36.964 5.047 7.324 0.000
## .V2 1.403 1.241 1.130 0.258
## .V3 10.209 1.400 7.292 0.000
## .V4 11.270 1.520 7.414 0.000
## .V5 3.996 1.192 3.353 0.001
## .V6 9.389 1.337 7.021 0.000
## .V7 9.575 1.353 7.076 0.000
## .V8 7.067 1.384 5.106 0.000
## .V9 11.736 2.438 4.814 0.000
## interc 112.909 13.915 8.114 0.000
## slope1 3.697 0.516 7.160 0.000
## slope2 2.949 0.435 6.782 0.000Spline Model-2
spline.model2 <- '
interc =~ 1*V1 + 1*V2 + 1*V3 + 1*V4 + 1*V5 + 1*V6 + 1*V7 + 1*V8 + 1*V9
slope1 =~ 0*V1 + 1*V2 + 2*V3 + 3*V4 + 3*V5 + 3*V6 + 3*V7 + 3*V8 + 3*V9
slope2 =~ 0*V1 + 0*V2 + 0*V3 + 0*V4 + 1*V5 + 2*V6 + 3*V7 + 4*V8 + 5*V9
'
spline.fit2 <- growth(spline.model2, sample.cov = covmat, sample.mean = smeans, sample.nobs = 140)
summary(spline.fit2, fit.measures = T)## lavaan 0.6.15 ended normally after 109 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 18
##
## Number of observations 140
##
## Model Test User Model:
##
## Test statistic 158.440
## Degrees of freedom 36
## P-value (Chi-square) 0.000
##
## Model Test Baseline Model:
##
## Test statistic 1702.962
## Degrees of freedom 36
## P-value 0.000
##
## User Model versus Baseline Model:
##
## Comparative Fit Index (CFI) 0.927
## Tucker-Lewis Index (TLI) 0.927
##
## Loglikelihood and Information Criteria:
##
## Loglikelihood user model (H0) -3724.429
## Loglikelihood unrestricted model (H1) -3645.210
##
## Akaike (AIC) 7484.859
## Bayesian (BIC) 7537.809
## Sample-size adjusted Bayesian (SABIC) 7480.859
##
## Root Mean Square Error of Approximation:
##
## RMSEA 0.156
## 90 Percent confidence interval - lower 0.132
## 90 Percent confidence interval - upper 0.181
## P-value H_0: RMSEA <= 0.050 0.000
## P-value H_0: RMSEA >= 0.080 1.000
##
## Standardized Root Mean Square Residual:
##
## SRMR 0.089
##
## Parameter Estimates:
##
## Standard errors Standard
## Information Expected
## Information saturated (h1) model Structured
##
## Latent Variables:
## Estimate Std.Err z-value P(>|z|)
## interc =~
## V1 1.000
## V2 1.000
## V3 1.000
## V4 1.000
## V5 1.000
## V6 1.000
## V7 1.000
## V8 1.000
## V9 1.000
## slope1 =~
## V1 0.000
## V2 1.000
## V3 2.000
## V4 3.000
## V5 3.000
## V6 3.000
## V7 3.000
## V8 3.000
## V9 3.000
## slope2 =~
## V1 0.000
## V2 0.000
## V3 0.000
## V4 0.000
## V5 1.000
## V6 2.000
## V7 3.000
## V8 4.000
## V9 5.000
##
## Covariances:
## Estimate Std.Err z-value P(>|z|)
## interc ~~
## slope1 -9.999 2.432 -4.112 0.000
## slope2 -7.849 1.671 -4.697 0.000
## slope1 ~~
## slope2 -0.363 0.374 -0.970 0.332
##
## Intercepts:
## Estimate Std.Err z-value P(>|z|)
## .V1 0.000
## .V2 0.000
## .V3 0.000
## .V4 0.000
## .V5 0.000
## .V6 0.000
## .V7 0.000
## .V8 0.000
## .V9 0.000
## interc 21.471 0.876 24.508 0.000
## slope1 3.781 0.205 18.429 0.000
## slope2 1.449 0.149 9.695 0.000
##
## Variances:
## Estimate Std.Err z-value P(>|z|)
## .V1 25.455 4.207 6.051 0.000
## .V2 4.277 1.350 3.169 0.002
## .V3 8.643 1.272 6.797 0.000
## .V4 6.259 1.376 4.548 0.000
## .V5 7.983 1.219 6.548 0.000
## .V6 11.116 1.500 7.411 0.000
## .V7 9.711 1.397 6.952 0.000
## .V8 6.363 1.353 4.703 0.000
## .V9 15.523 2.682 5.788 0.000
## interc 100.209 12.905 7.765 0.000
## slope1 4.339 0.725 5.984 0.000
## slope2 2.676 0.376 7.119 0.000Spline Model-3
spline.model3 <- '
interc =~ 1*V1 + 1*V2 + 1*V3 + 1*V4 + 1*V5 + 1*V6 + 1*V7 + 1*V8 + 1*V9
slope1 =~ 0*V1 + 1*V2 + 2*V3 + 2*V4 + 2*V5 + 2*V6 + 2*V7 + 2*V8 + 2*V9
slope2 =~ 0*V1 + 0*V2 + 0*V3 + 1*V4 + 2*V5 + 3*V6 + 4*V7 + 5*V8 + 6*V9
'
spline.fit3 <- growth(spline.model3, sample.cov = covmat, sample.mean = smeans, sample.nobs = 140)
summary(spline.fit3, fit.measures = T)## lavaan 0.6.15 ended normally after 103 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 18
##
## Number of observations 140
##
## Model Test User Model:
##
## Test statistic 205.005
## Degrees of freedom 36
## P-value (Chi-square) 0.000
##
## Model Test Baseline Model:
##
## Test statistic 1702.962
## Degrees of freedom 36
## P-value 0.000
##
## User Model versus Baseline Model:
##
## Comparative Fit Index (CFI) 0.899
## Tucker-Lewis Index (TLI) 0.899
##
## Loglikelihood and Information Criteria:
##
## Loglikelihood user model (H0) -3747.712
## Loglikelihood unrestricted model (H1) -3645.210
##
## Akaike (AIC) 7531.425
## Bayesian (BIC) 7584.374
## Sample-size adjusted Bayesian (SABIC) 7527.425
##
## Root Mean Square Error of Approximation:
##
## RMSEA 0.183
## 90 Percent confidence interval - lower 0.159
## 90 Percent confidence interval - upper 0.208
## P-value H_0: RMSEA <= 0.050 0.000
## P-value H_0: RMSEA >= 0.080 1.000
##
## Standardized Root Mean Square Residual:
##
## SRMR 0.067
##
## Parameter Estimates:
##
## Standard errors Standard
## Information Expected
## Information saturated (h1) model Structured
##
## Latent Variables:
## Estimate Std.Err z-value P(>|z|)
## interc =~
## V1 1.000
## V2 1.000
## V3 1.000
## V4 1.000
## V5 1.000
## V6 1.000
## V7 1.000
## V8 1.000
## V9 1.000
## slope1 =~
## V1 0.000
## V2 1.000
## V3 2.000
## V4 2.000
## V5 2.000
## V6 2.000
## V7 2.000
## V8 2.000
## V9 2.000
## slope2 =~
## V1 0.000
## V2 0.000
## V3 0.000
## V4 1.000
## V5 2.000
## V6 3.000
## V7 4.000
## V8 5.000
## V9 6.000
##
## Covariances:
## Estimate Std.Err z-value P(>|z|)
## interc ~~
## slope1 -7.769 3.312 -2.346 0.019
## slope2 -6.923 1.424 -4.862 0.000
## slope1 ~~
## slope2 -1.240 0.500 -2.480 0.013
##
## Intercepts:
## Estimate Std.Err z-value P(>|z|)
## .V1 0.000
## .V2 0.000
## .V3 0.000
## .V4 0.000
## .V5 0.000
## .V6 0.000
## .V7 0.000
## .V8 0.000
## .V9 0.000
## interc 20.240 0.819 24.716 0.000
## slope1 5.085 0.295 17.250 0.000
## slope2 1.683 0.135 12.507 0.000
##
## Variances:
## Estimate Std.Err z-value P(>|z|)
## .V1 9.866 3.834 2.573 0.010
## .V2 8.111 1.520 5.337 0.000
## .V3 10.681 1.942 5.498 0.000
## .V4 8.502 1.391 6.111 0.000
## .V5 9.845 1.386 7.105 0.000
## .V6 11.624 1.567 7.417 0.000
## .V7 9.474 1.398 6.776 0.000
## .V8 6.857 1.404 4.884 0.000
## .V9 18.589 2.950 6.301 0.000
## interc 86.088 11.453 7.517 0.000
## slope1 8.661 1.647 5.260 0.000
## slope2 2.174 0.304 7.150 0.000Spline Model-4
spline.model4 <- '
interc =~ 1*V1 + 1*V2 + 1*V3 + 1*V4 + 1*V5 + 1*V6 + 1*V7 + 1*V8 + 1*V9
slope1 =~ 0*V1 + 1*V2 + 2*V3 + 2*V4 + 2*V5 + 2*V6 + 2*V7 + 2*V8 + 2*V9
slope2 =~ 0*V1 + 0*V2 + 0*V3 + 1*V4 + 2*V5 + 3*V6 + 4*V7 + 5*V8 + 6*V9
slope3 =~ 0*V1 + 0*V2 + 0*V3 + 0*V4 + 0*V5 + 0*V6 + 1*V7 + 2*V8 + 3*V9
'
spline.fit4 <- growth(spline.model4, sample.cov = covmat, sample.mean = smeans, sample.nobs = 140)
summary(spline.fit4, fit.measures = T)## lavaan 0.6.15 ended normally after 144 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 23
##
## Number of observations 140
##
## Model Test User Model:
##
## Test statistic 100.459
## Degrees of freedom 31
## P-value (Chi-square) 0.000
##
## Model Test Baseline Model:
##
## Test statistic 1702.962
## Degrees of freedom 36
## P-value 0.000
##
## User Model versus Baseline Model:
##
## Comparative Fit Index (CFI) 0.958
## Tucker-Lewis Index (TLI) 0.952
##
## Loglikelihood and Information Criteria:
##
## Loglikelihood user model (H0) -3695.439
## Loglikelihood unrestricted model (H1) -3645.210
##
## Akaike (AIC) 7436.878
## Bayesian (BIC) 7504.536
## Sample-size adjusted Bayesian (SABIC) 7431.767
##
## Root Mean Square Error of Approximation:
##
## RMSEA 0.127
## 90 Percent confidence interval - lower 0.099
## 90 Percent confidence interval - upper 0.155
## P-value H_0: RMSEA <= 0.050 0.000
## P-value H_0: RMSEA >= 0.080 0.997
##
## Standardized Root Mean Square Residual:
##
## SRMR 0.045
##
## Parameter Estimates:
##
## Standard errors Standard
## Information Expected
## Information saturated (h1) model Structured
##
## Latent Variables:
## Estimate Std.Err z-value P(>|z|)
## interc =~
## V1 1.000
## V2 1.000
## V3 1.000
## V4 1.000
## V5 1.000
## V6 1.000
## V7 1.000
## V8 1.000
## V9 1.000
## slope1 =~
## V1 0.000
## V2 1.000
## V3 2.000
## V4 2.000
## V5 2.000
## V6 2.000
## V7 2.000
## V8 2.000
## V9 2.000
## slope2 =~
## V1 0.000
## V2 0.000
## V3 0.000
## V4 1.000
## V5 2.000
## V6 3.000
## V7 4.000
## V8 5.000
## V9 6.000
## slope3 =~
## V1 0.000
## V2 0.000
## V3 0.000
## V4 0.000
## V5 0.000
## V6 0.000
## V7 1.000
## V8 2.000
## V9 3.000
##
## Covariances:
## Estimate Std.Err z-value P(>|z|)
## interc ~~
## slope1 -3.053 2.796 -1.092 0.275
## slope2 -11.537 2.090 -5.520 0.000
## slope3 9.383 2.776 3.381 0.001
## slope1 ~~
## slope2 -0.788 0.678 -1.162 0.245
## slope3 -0.627 0.931 -0.674 0.500
## slope2 ~~
## slope3 -3.988 0.822 -4.851 0.000
##
## Intercepts:
## Estimate Std.Err z-value P(>|z|)
## .V1 0.000
## .V2 0.000
## .V3 0.000
## .V4 0.000
## .V5 0.000
## .V6 0.000
## .V7 0.000
## .V8 0.000
## .V9 0.000
## interc 20.653 0.815 25.332 0.000
## slope1 4.497 0.266 16.880 0.000
## slope2 2.199 0.193 11.400 0.000
## slope3 -1.135 0.276 -4.109 0.000
##
## Variances:
## Estimate Std.Err z-value P(>|z|)
## .V1 16.174 3.680 4.396 0.000
## .V2 5.626 1.240 4.538 0.000
## .V3 4.327 1.466 2.951 0.003
## .V4 10.102 1.427 7.079 0.000
## .V5 7.564 1.149 6.584 0.000
## .V6 7.130 1.513 4.712 0.000
## .V7 8.861 1.318 6.723 0.000
## .V8 7.794 1.408 5.537 0.000
## .V9 9.037 2.474 3.653 0.000
## interc 83.075 11.205 7.414 0.000
## slope1 5.939 1.351 4.396 0.000
## slope2 4.256 0.641 6.634 0.000
## slope3 7.016 1.356 5.173 0.000Unspecified Loadings
model5 <- '
interc =~ 1*V1 + 1*V2 + 1*V3 + 1*V4 + 1*V5 + 1*V6 + 1*V7 + 1*V8 + 1*V9
slope =~ 0*V1 + 1*V2 + NA*V3 + NA*V4 + NA*V5 + NA*V6 + NA*V7 + NA*V8 + NA*V9
'
fit5 <- growth(model5, sample.cov = covmat, sample.mean = smeans, sample.nobs = 140)
summary(fit5, fit.measures = T)## lavaan 0.6.15 ended normally after 96 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 21
##
## Number of observations 140
##
## Model Test User Model:
##
## Test statistic 276.817
## Degrees of freedom 33
## P-value (Chi-square) 0.000
##
## Model Test Baseline Model:
##
## Test statistic 1702.962
## Degrees of freedom 36
## P-value 0.000
##
## User Model versus Baseline Model:
##
## Comparative Fit Index (CFI) 0.854
## Tucker-Lewis Index (TLI) 0.840
##
## Loglikelihood and Information Criteria:
##
## Loglikelihood user model (H0) -3783.618
## Loglikelihood unrestricted model (H1) -3645.210
##
## Akaike (AIC) 7609.236
## Bayesian (BIC) 7671.011
## Sample-size adjusted Bayesian (SABIC) 7604.570
##
## Root Mean Square Error of Approximation:
##
## RMSEA 0.230
## 90 Percent confidence interval - lower 0.205
## 90 Percent confidence interval - upper 0.255
## P-value H_0: RMSEA <= 0.050 0.000
## P-value H_0: RMSEA >= 0.080 1.000
##
## Standardized Root Mean Square Residual:
##
## SRMR 0.134
##
## Parameter Estimates:
##
## Standard errors Standard
## Information Expected
## Information saturated (h1) model Structured
##
## Latent Variables:
## Estimate Std.Err z-value P(>|z|)
## interc =~
## V1 1.000
## V2 1.000
## V3 1.000
## V4 1.000
## V5 1.000
## V6 1.000
## V7 1.000
## V8 1.000
## V9 1.000
## slope =~
## V1 0.000
## V2 1.000
## V3 1.872 0.144 13.018 0.000
## V4 2.635 0.236 11.177 0.000
## V5 3.227 0.304 10.623 0.000
## V6 3.657 0.355 10.287 0.000
## V7 4.245 0.426 9.958 0.000
## V8 4.499 0.459 9.798 0.000
## V9 4.499 0.463 9.721 0.000
##
## Covariances:
## Estimate Std.Err z-value P(>|z|)
## interc ~~
## slope -17.920 3.803 -4.712 0.000
##
## Intercepts:
## Estimate Std.Err z-value P(>|z|)
## .V1 0.000
## .V2 0.000
## .V3 0.000
## .V4 0.000
## .V5 0.000
## .V6 0.000
## .V7 0.000
## .V8 0.000
## .V9 0.000
## interc 21.658 1.042 20.777 0.000
## slope 3.848 0.518 7.423 0.000
##
## Variances:
## Estimate Std.Err z-value P(>|z|)
## .V1 36.932 5.210 7.089 0.000
## .V2 3.583 1.410 2.541 0.011
## .V3 10.788 1.539 7.008 0.000
## .V4 16.544 2.122 7.797 0.000
## .V5 12.823 1.696 7.562 0.000
## .V6 11.437 1.580 7.239 0.000
## .V7 7.488 1.284 5.834 0.000
## .V8 10.658 1.702 6.263 0.000
## .V9 19.512 2.698 7.233 0.000
## interc 121.309 15.592 7.780 0.000
## slope 4.444 1.254 3.544 0.000© Copyright 2024 @Yi Feng and @Gregory R. Hancock.