Panel Models
In this tutorial, we are going to use lavaan for panel
models.
Load the pacakges
library(lavaan)
library(semPlot)Example: Panel Model for Read
Read the data
setwd(mypath) # change it to the path of your own data folder
dat <- read.delim("panel_data.txt", sep = "\t", header = F)
colnames(dat) <- c(paste0("read", 1:3), paste0("list", 1:3), paste0("speak", 1:3), paste0("write", 1:3))
# check the data
str(dat)## 'data.frame': 170 obs. of 12 variables:
## $ read1 : int 22 19 24 21 24 17 14 30 19 11 ...
## $ read2 : int 24 19 26 24 26 21 19 29 23 6 ...
## $ read3 : int 23 17 30 27 28 20 20 29 23 13 ...
## $ list1 : int 28 18 27 18 29 18 14 27 13 15 ...
## $ list2 : int 30 16 30 20 28 16 7 18 10 8 ...
## $ list3 : int 29 17 27 18 26 17 10 24 17 19 ...
## $ speak1: int 23 19 24 22 17 14 14 23 17 23 ...
## $ speak2: int 23 18 24 17 18 17 10 22 17 22 ...
## $ speak3: int 23 18 29 17 19 15 10 23 18 24 ...
## $ write1: int 25 17 21 18 20 21 14 22 17 17 ...
## $ write2: int 22 20 24 21 20 17 18 25 20 18 ...
## $ write3: int 21 25 27 22 22 21 18 22 17 18 ...summary(dat)## read1 read2 read3 list1
## Min. : 0.00 Min. : 3.00 Min. : 4.00 Min. : 1.00
## 1st Qu.:13.00 1st Qu.:14.00 1st Qu.:16.00 1st Qu.:13.00
## Median :17.00 Median :19.00 Median :20.00 Median :17.00
## Mean :17.38 Mean :18.55 Mean :19.55 Mean :16.56
## 3rd Qu.:22.00 3rd Qu.:23.00 3rd Qu.:23.00 3rd Qu.:21.00
## Max. :30.00 Max. :30.00 Max. :30.00 Max. :30.00
## list2 list3 speak1 speak2
## Min. : 1.00 Min. : 1.00 Min. : 0.00 Min. : 5.00
## 1st Qu.:14.00 1st Qu.:15.00 1st Qu.:15.00 1st Qu.:17.00
## Median :18.00 Median :19.00 Median :18.00 Median :18.00
## Mean :17.63 Mean :18.58 Mean :17.96 Mean :18.73
## 3rd Qu.:21.75 3rd Qu.:23.00 3rd Qu.:20.00 3rd Qu.:22.00
## Max. :30.00 Max. :30.00 Max. :28.00 Max. :27.00
## speak3 write1 write2 write3
## Min. :10.00 Min. : 7.00 Min. : 8.00 Min. : 7.00
## 1st Qu.:17.00 1st Qu.:15.00 1st Qu.:17.00 1st Qu.:18.00
## Median :19.00 Median :20.00 Median :20.00 Median :20.50
## Mean :19.24 Mean :18.59 Mean :19.58 Mean :20.11
## 3rd Qu.:22.00 3rd Qu.:21.00 3rd Qu.:22.00 3rd Qu.:22.00
## Max. :29.00 Max. :30.00 Max. :28.00 Max. :29.00Fit the panel model to the data
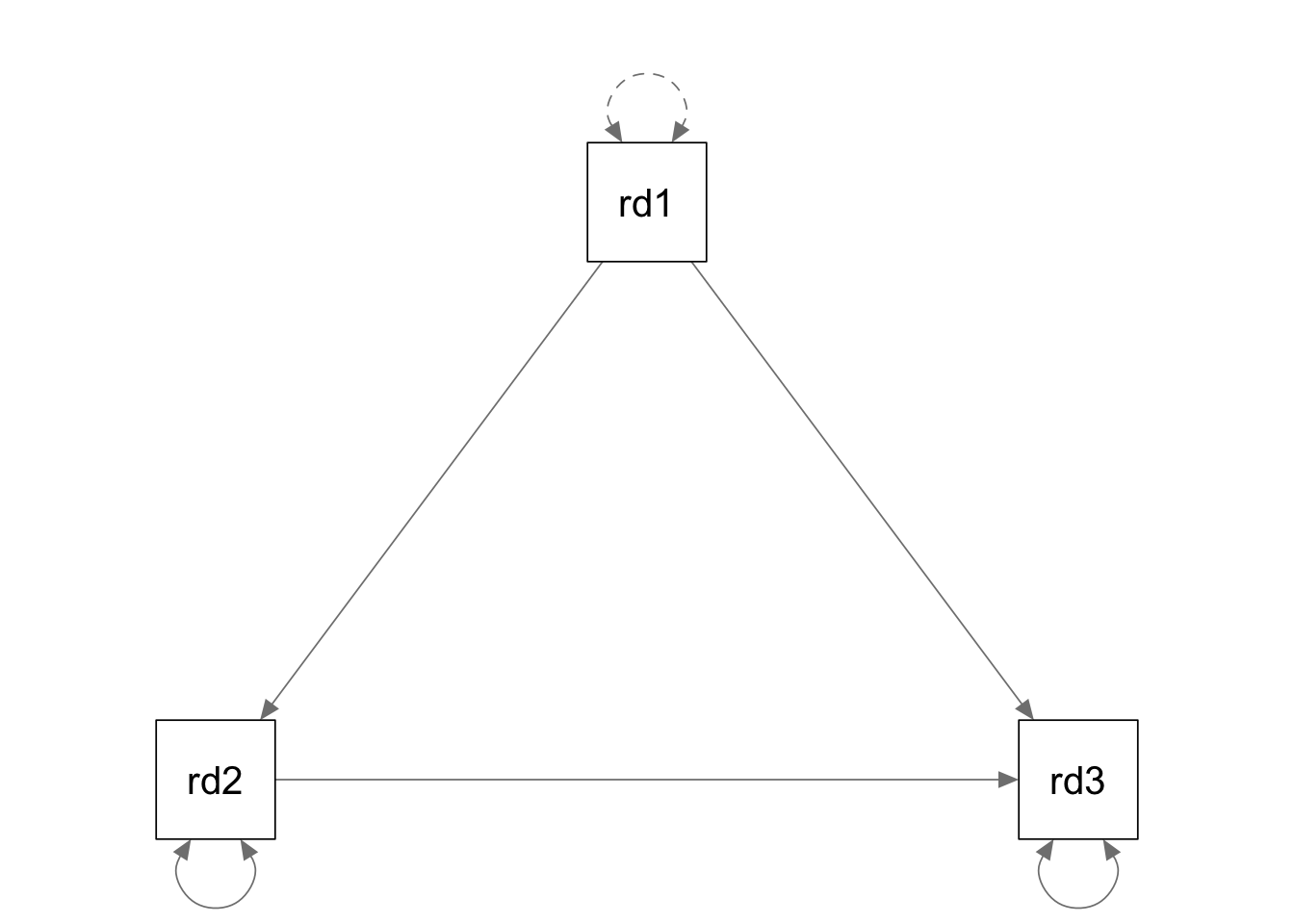
rd.model <- '
read2 ~ a*read1
read3 ~ a*read2 + read1
'
rd.fit <- sem(rd.model, dat)
summary(rd.fit, fit.measures = T, standardized = T)## lavaan 0.6.15 ended normally after 10 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 5
## Number of equality constraints 1
##
## Number of observations 170
##
## Model Test User Model:
##
## Test statistic 9.372
## Degrees of freedom 1
## P-value (Chi-square) 0.002
##
## Model Test Baseline Model:
##
## Test statistic 306.585
## Degrees of freedom 3
## P-value 0.000
##
## User Model versus Baseline Model:
##
## Comparative Fit Index (CFI) 0.972
## Tucker-Lewis Index (TLI) 0.917
##
## Loglikelihood and Information Criteria:
##
## Loglikelihood user model (H0) -934.038
## Loglikelihood unrestricted model (H1) -929.352
##
## Akaike (AIC) 1876.076
## Bayesian (BIC) 1888.619
## Sample-size adjusted Bayesian (SABIC) 1875.954
##
## Root Mean Square Error of Approximation:
##
## RMSEA 0.222
## 90 Percent confidence interval - lower 0.109
## 90 Percent confidence interval - upper 0.361
## P-value H_0: RMSEA <= 0.050 0.008
## P-value H_0: RMSEA >= 0.080 0.978
##
## Standardized Root Mean Square Residual:
##
## SRMR 0.070
##
## Parameter Estimates:
##
## Standard errors Standard
## Information Expected
## Information saturated (h1) model Structured
##
## Regressions:
## Estimate Std.Err z-value P(>|z|) Std.lv Std.all
## read2 ~
## read1 (a) 0.612 0.039 15.666 0.000 0.612 0.689
## read3 ~
## read2 (a) 0.612 0.039 15.666 0.000 0.612 0.618
## read1 0.201 0.048 4.216 0.000 0.201 0.228
##
## Variances:
## Estimate Std.Err z-value P(>|z|) Std.lv Std.all
## .read2 17.084 1.853 9.220 0.000 17.084 0.525
## .read3 11.879 1.288 9.220 0.000 11.879 0.372Example: Panel Model for Speak
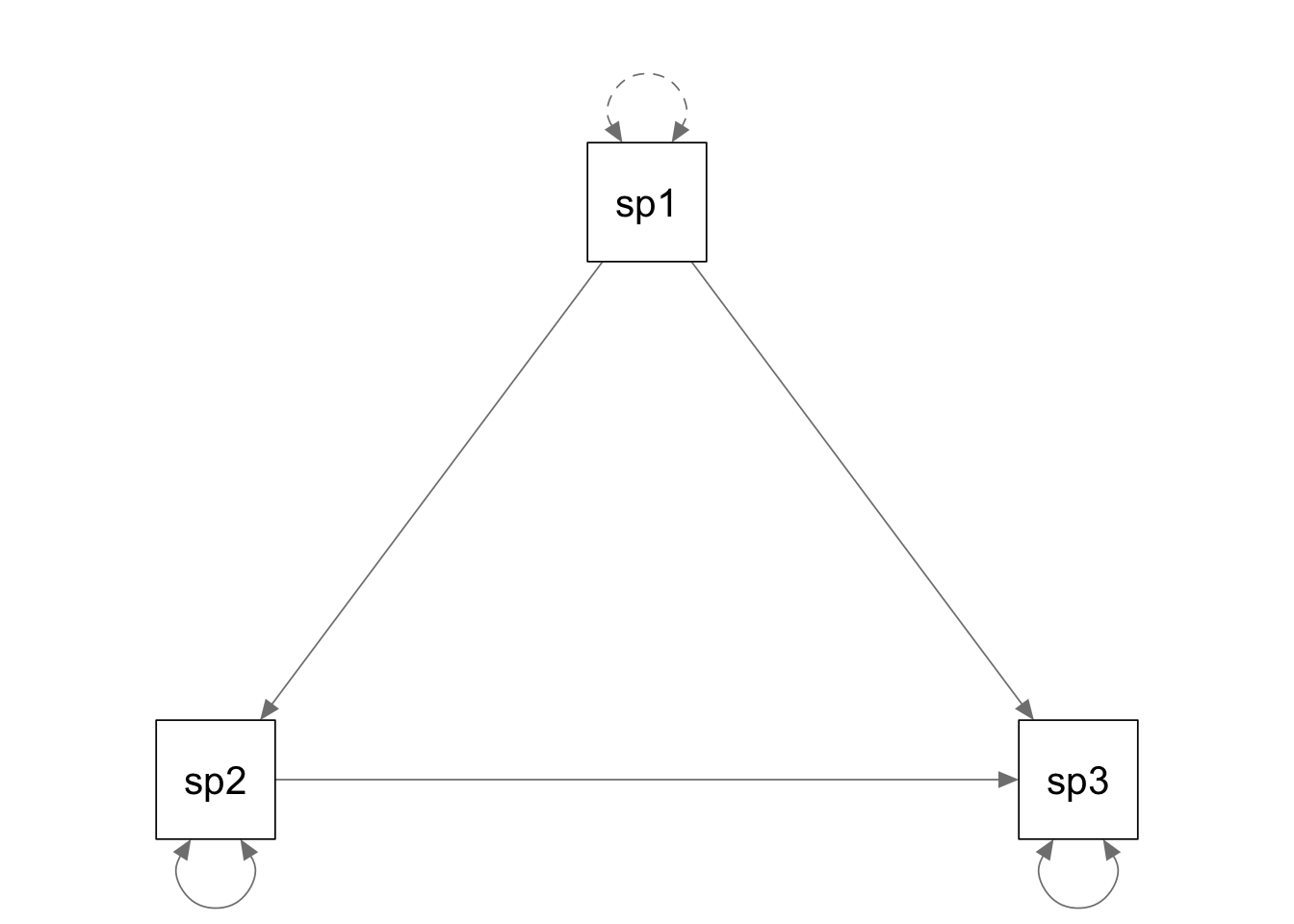
spk.model <- '
speak2 ~ a*speak1
speak3 ~ a*speak2 + speak1
'
spk.fit <- sem(spk.model, dat)
summary(spk.fit, fit.measures = T, standardized = T)## lavaan 0.6.15 ended normally after 11 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 5
## Number of equality constraints 1
##
## Number of observations 170
##
## Model Test User Model:
##
## Test statistic 5.086
## Degrees of freedom 1
## P-value (Chi-square) 0.024
##
## Model Test Baseline Model:
##
## Test statistic 362.145
## Degrees of freedom 3
## P-value 0.000
##
## User Model versus Baseline Model:
##
## Comparative Fit Index (CFI) 0.989
## Tucker-Lewis Index (TLI) 0.966
##
## Loglikelihood and Information Criteria:
##
## Loglikelihood user model (H0) -755.917
## Loglikelihood unrestricted model (H1) -753.374
##
## Akaike (AIC) 1519.835
## Bayesian (BIC) 1532.378
## Sample-size adjusted Bayesian (SABIC) 1519.713
##
## Root Mean Square Error of Approximation:
##
## RMSEA 0.155
## 90 Percent confidence interval - lower 0.045
## 90 Percent confidence interval - upper 0.299
## P-value H_0: RMSEA <= 0.050 0.056
## P-value H_0: RMSEA >= 0.080 0.887
##
## Standardized Root Mean Square Residual:
##
## SRMR 0.051
##
## Parameter Estimates:
##
## Standard errors Standard
## Information Expected
## Information saturated (h1) model Structured
##
## Regressions:
## Estimate Std.Err z-value P(>|z|) Std.lv Std.all
## speak2 ~
## speak1 (a) 0.679 0.037 18.143 0.000 0.679 0.746
## speak3 ~
## speak2 (a) 0.679 0.037 18.143 0.000 0.679 0.682
## speak1 0.174 0.046 3.786 0.000 0.174 0.192
##
## Variances:
## Estimate Std.Err z-value P(>|z|) Std.lv Std.all
## .speak2 6.080 0.659 9.220 0.000 6.080 0.444
## .speak3 4.106 0.445 9.220 0.000 4.106 0.303Example: Panel Model for Read and Speak
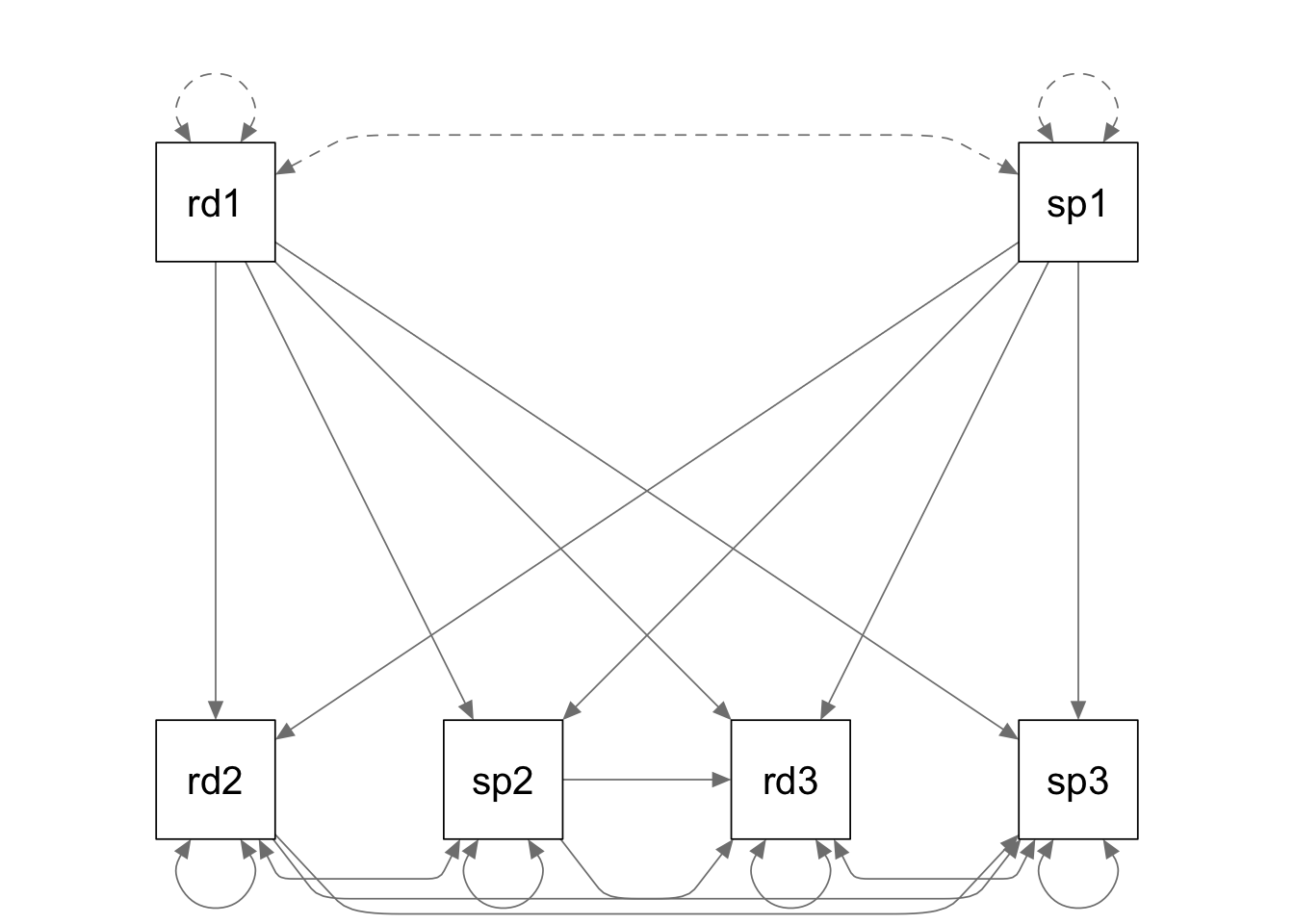
rdspk.model <- '
read2 ~ read1 + speak1
speak2 ~ speak1 + read1
read3 ~ read1 + read2 + speak1 + speak2
speak3 ~ speak1 + speak2 + read1 + read2;
read2 ~~ speak2
read3 ~~ speak3
'
rdspk.fit <- sem(rdspk.model, dat)
summary(rdspk.fit, fit.measures = T, standardized = T)## lavaan 0.6.15 ended normally after 22 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 18
##
## Number of observations 170
##
## Model Test User Model:
##
## Test statistic 0.000
## Degrees of freedom 0
##
## Model Test Baseline Model:
##
## Test statistic 679.923
## Degrees of freedom 14
## P-value 0.000
##
## User Model versus Baseline Model:
##
## Comparative Fit Index (CFI) 1.000
## Tucker-Lewis Index (TLI) 1.000
##
## Loglikelihood and Information Criteria:
##
## Loglikelihood user model (H0) -1677.130
## Loglikelihood unrestricted model (H1) -1677.130
##
## Akaike (AIC) 3390.260
## Bayesian (BIC) 3446.705
## Sample-size adjusted Bayesian (SABIC) 3389.710
##
## Root Mean Square Error of Approximation:
##
## RMSEA 0.000
## 90 Percent confidence interval - lower 0.000
## 90 Percent confidence interval - upper 0.000
## P-value H_0: RMSEA <= 0.050 NA
## P-value H_0: RMSEA >= 0.080 NA
##
## Standardized Root Mean Square Residual:
##
## SRMR 0.000
##
## Parameter Estimates:
##
## Standard errors Standard
## Information Expected
## Information saturated (h1) model Structured
##
## Regressions:
## Estimate Std.Err z-value P(>|z|) Std.lv Std.all
## read2 ~
## read1 0.674 0.051 13.216 0.000 0.674 0.709
## speak1 0.151 0.080 1.879 0.060 0.151 0.101
## speak2 ~
## speak1 0.706 0.048 14.717 0.000 0.706 0.739
## read1 0.071 0.030 2.332 0.020 0.071 0.117
## read3 ~
## read1 0.303 0.061 4.995 0.000 0.303 0.347
## read2 0.447 0.064 6.975 0.000 0.447 0.487
## speak1 0.039 0.101 0.389 0.697 0.039 0.029
## speak2 0.028 0.108 0.258 0.797 0.028 0.019
## speak3 ~
## speak1 0.258 0.060 4.285 0.000 0.258 0.285
## speak2 0.562 0.064 8.799 0.000 0.562 0.594
## read1 -0.005 0.036 -0.152 0.879 -0.005 -0.010
## read2 0.011 0.038 0.284 0.777 0.011 0.018
##
## Covariances:
## Estimate Std.Err z-value P(>|z|) Std.lv Std.all
## .read2 ~~
## .speak2 0.679 0.752 0.903 0.366 0.679 0.069
## .read3 ~~
## .speak3 0.409 0.521 0.785 0.433 0.409 0.060
##
## Variances:
## Estimate Std.Err z-value P(>|z|) Std.lv Std.all
## .read2 16.401 1.779 9.220 0.000 16.401 0.442
## .speak2 5.830 0.632 9.220 0.000 5.830 0.386
## .read3 11.417 1.238 9.220 0.000 11.417 0.364
## .speak3 4.025 0.437 9.220 0.000 4.025 0.298Example: Panel Model for English
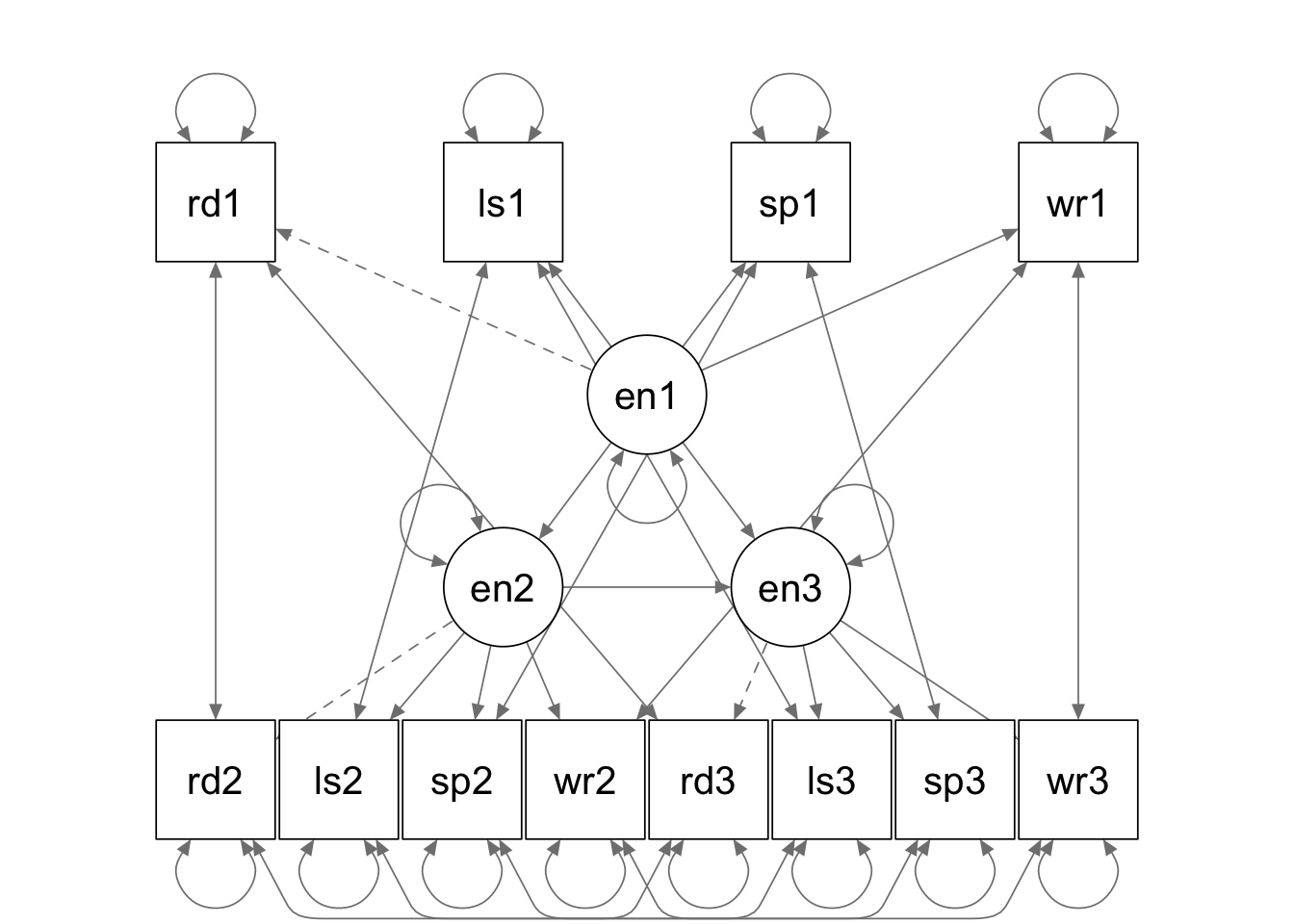
eng.model <- '
english1 =~ 1*read1 + a*list1 + b*speak1 + c*write1
english2 =~ 1*read2 + a*list2 + b*speak2 + c*write2
english3 =~ 1*read3 + a*list3 + b*speak3 + c*write3
english2 ~ english1
english3 ~ english2 + english1
read1 ~~ read2 + read3
read2 ~~ read3
list1 ~~ list2 + list3
list2 ~~ list3
speak1 ~~ speak2 + speak3
speak2 ~~ speak3
write1 ~~ write2 + write3
write2 ~~ write3
'
eng.fit <- sem(eng.model, dat, estimator = "MLR")
summary(eng.fit, fit.measures = T, standardized = T)## lavaan 0.6.15 ended normally after 133 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 39
## Number of equality constraints 6
##
## Number of observations 170
##
## Model Test User Model:
## Standard Scaled
## Test Statistic 106.529 107.877
## Degrees of freedom 45 45
## P-value (Chi-square) 0.000 0.000
## Scaling correction factor 0.988
## Yuan-Bentler correction (Mplus variant)
##
## Model Test Baseline Model:
##
## Test statistic 1711.627 1475.426
## Degrees of freedom 66 66
## P-value 0.000 0.000
## Scaling correction factor 1.160
##
## User Model versus Baseline Model:
##
## Comparative Fit Index (CFI) 0.963 0.955
## Tucker-Lewis Index (TLI) 0.945 0.935
##
## Robust Comparative Fit Index (CFI) 0.962
## Robust Tucker-Lewis Index (TLI) 0.944
##
## Loglikelihood and Information Criteria:
##
## Loglikelihood user model (H0) -5351.557 -5351.557
## Scaling correction factor 1.130
## for the MLR correction
## Loglikelihood unrestricted model (H1) -5298.293 -5298.293
## Scaling correction factor 1.135
## for the MLR correction
##
## Akaike (AIC) 10769.115 10769.115
## Bayesian (BIC) 10872.596 10872.596
## Sample-size adjusted Bayesian (SABIC) 10768.106 10768.106
##
## Root Mean Square Error of Approximation:
##
## RMSEA 0.090 0.091
## 90 Percent confidence interval - lower 0.068 0.069
## 90 Percent confidence interval - upper 0.112 0.113
## P-value H_0: RMSEA <= 0.050 0.002 0.002
## P-value H_0: RMSEA >= 0.080 0.779 0.799
##
## Robust RMSEA 0.090
## 90 Percent confidence interval - lower 0.068
## 90 Percent confidence interval - upper 0.112
## P-value H_0: Robust RMSEA <= 0.050 0.002
## P-value H_0: Robust RMSEA >= 0.080 0.789
##
## Standardized Root Mean Square Residual:
##
## SRMR 0.069 0.069
##
## Parameter Estimates:
##
## Standard errors Sandwich
## Information bread Observed
## Observed information based on Hessian
##
## Latent Variables:
## Estimate Std.Err z-value P(>|z|) Std.lv Std.all
## english1 =~
## read1 1.000 4.249 0.655
## list1 (a) 1.165 0.103 11.361 0.000 4.950 0.797
## speak1 (b) 0.675 0.091 7.392 0.000 2.868 0.708
## write1 (c) 0.864 0.078 11.062 0.000 3.671 0.843
## english2 =~
## read2 1.000 3.914 0.660
## list2 (a) 1.165 0.103 11.361 0.000 4.560 0.710
## speak2 (b) 0.675 0.091 7.392 0.000 2.642 0.700
## write2 (c) 0.864 0.078 11.062 0.000 3.382 0.801
## english3 =~
## read3 1.000 3.893 0.692
## list3 (a) 1.165 0.103 11.361 0.000 4.535 0.785
## speak3 (b) 0.675 0.091 7.392 0.000 2.628 0.705
## write3 (c) 0.864 0.078 11.062 0.000 3.363 0.821
##
## Regressions:
## Estimate Std.Err z-value P(>|z|) Std.lv Std.all
## english2 ~
## english1 0.880 0.051 17.414 0.000 0.955 0.955
## english3 ~
## english2 1.246 0.313 3.975 0.000 1.253 1.253
## english1 -0.257 0.287 -0.895 0.371 -0.280 -0.280
##
## Covariances:
## Estimate Std.Err z-value P(>|z|) Std.lv Std.all
## .read1 ~~
## .read2 12.128 1.758 6.900 0.000 12.128 0.556
## .read3 11.272 1.841 6.124 0.000 11.272 0.565
## .read2 ~~
## .read3 10.220 1.719 5.945 0.000 10.220 0.565
## .list1 ~~
## .list2 5.653 2.058 2.747 0.006 5.653 0.333
## .list3 3.795 1.756 2.161 0.031 3.795 0.283
## .list2 ~~
## .list3 5.690 2.255 2.523 0.012 5.690 0.352
## .speak1 ~~
## .speak2 4.498 0.785 5.731 0.000 4.498 0.583
## .speak3 4.302 0.756 5.694 0.000 4.302 0.568
## .speak2 ~~
## .speak3 4.585 0.789 5.813 0.000 4.585 0.644
## .write1 ~~
## .write2 1.552 0.812 1.911 0.056 1.552 0.262
## .write3 1.676 0.744 2.253 0.024 1.676 0.306
## .write2 ~~
## .write3 2.371 0.882 2.688 0.007 2.371 0.400
##
## Variances:
## Estimate Std.Err z-value P(>|z|) Std.lv Std.all
## .read1 24.058 2.952 8.149 0.000 24.058 0.571
## .list1 14.084 2.461 5.723 0.000 14.084 0.365
## .speak1 8.207 1.524 5.386 0.000 8.207 0.499
## .write1 5.478 1.034 5.296 0.000 5.478 0.289
## .read2 19.805 2.401 8.249 0.000 19.805 0.564
## .list2 20.422 3.098 6.592 0.000 20.422 0.496
## .speak2 7.261 0.935 7.767 0.000 7.261 0.510
## .write2 6.391 0.999 6.400 0.000 6.391 0.358
## .read3 16.528 2.103 7.859 0.000 16.528 0.522
## .list3 12.810 2.129 6.017 0.000 12.810 0.384
## .speak3 6.987 0.868 8.048 0.000 6.987 0.503
## .write3 5.484 1.091 5.027 0.000 5.484 0.327
## english1 18.055 3.316 5.444 0.000 1.000 1.000
## .english2 1.336 0.620 2.155 0.031 0.087 0.087
## .english3 0.360 0.481 0.747 0.455 0.024 0.024© Copyright 2024 @Yi Feng and @Gregory R. Hancock.