EDMS 657-R Tutorial: MANOVA
Yi Feng & Gregory R. Hancock
University of Maryland, College ParkIn this document we supply three simple examples of MANOVA using R. This document serves as a supplementary material for EDMS 657-Exploratory Latent and Composite Variable Methods, taught by Dr. Gregory R. Hancock at University of Maryland.
Before we get started, let’s install all the packages we are going to use in this demo.
install.packages("tidyverse")
install.packages("ggplot2")
install.packages("gridExtra")
install.packages("heplots")
install.packages("agricolae")
install.packages("candisc")
install.packages("MASS")
install.packages("Hmisc")
devtools::install_github("YiFengEDMS/EDMS657Data")
Example 1: Two-group MANOVA / Hoteling’s \(T^2\)
Research Context
Middle school students are randomly assigned to an educational treatment or a control group. After receiving their treatment or control experience, students are evaluated in terms of cognitive and affective outcomes together. We are interested in testing whether there is any difference between the treatment/control groups regarding the set of outcomes.
Step 1: Read and examine the data
Load the data into the R session.
library(EDMS657Data)
data(Hotellings_T)Take a look at the data
library(tidyverse)
#View(Hotellings_T)
glimpse(Hotellings_T)## Rows: 30
## Columns: 3
## $ treat <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2…
## $ attitude <dbl> 4, 5, 6, 6, 7, 8, 8, 9, 10, 10, 11, 11, 12, 12, 13, 8, …
## $ know <dbl> 14, 14, 16, 20, 17, 15, 18, 13, 15, 21, 11, 12, 15, 24,…str(Hotellings_T)## 'data.frame': 30 obs. of 3 variables:
## $ treat : num 1 1 1 1 1 1 1 1 1 1 ...
## ..- attr(*, "value.labels")= Named chr [1:2] "2" "1"
## .. ..- attr(*, "names")= chr [1:2] "control" "experimental"
## $ attitude: num 4 5 6 6 7 8 8 9 10 10 ...
## $ know : num 14 14 16 20 17 15 18 13 15 21 ...
## - attr(*, "variable.labels")= Named chr [1:3] "treatment condition" "attitude toward people with HIV/AIDS" "HIV/AIDS knowledge test"
## ..- attr(*, "names")= chr [1:3] "treat" "attitude" "know"
## - attr(*, "codepage")= int 65001head(Hotellings_T)## treat attitude know
## 1 1 4 14
## 2 1 5 14
## 3 1 6 16
## 4 1 6 20
## 5 1 7 17
## 6 1 8 15- Note that “treatment” should be a categorical variable.
Hotellings_T <- Hotellings_T %>%
mutate(treat = factor(treat, labels = c("experimental", "control")))Step 2: Explore the data with descriptive statistics and plots
Descriptive statistics
summary(Hotellings_T)## treat attitude know
## experimental:15 Min. : 4.00 Min. :10.00
## control :15 1st Qu.: 8.25 1st Qu.:13.25
## Median :11.50 Median :15.00
## Mean :11.47 Mean :15.43
## 3rd Qu.:14.00 3rd Qu.:17.00
## Max. :19.00 Max. :24.00table(Hotellings_T$treat)##
## experimental control
## 15 15You can get the within-group means:
Hotellings_T %>%
group_by(treat) %>%
summarize(mean_att = mean(attitude),
mean_know = mean(know))## # A tibble: 2 × 3
## treat mean_att mean_know
## <fct> <dbl> <dbl>
## 1 experimental 8.8 16.2
## 2 control 14.1 14.7You can get the within-group variances:
Hotellings_T %>%
group_by(treat) %>%
summarize(var_att = var(attitude),
var_know = var(know))## # A tibble: 2 × 3
## treat var_att var_know
## <fct> <dbl> <dbl>
## 1 experimental 7.74 12.5
## 2 control 10.4 6.95Or alternatively, examine group statistics altogether:
psych::describeBy(attitude + know ~ treat, data = Hotellings_T)##
## Descriptive statistics by group
## treat: experimental
## vars n mean sd median trimmed mad min max range skew
## attitude 1 15 8.8 2.78 9 8.85 2.97 4 13 9 -0.16
## know 2 15 16.2 3.53 15 16.00 2.97 11 24 13 0.57
## kurtosis se
## attitude -1.39 0.72
## know -0.57 0.91
## -------------------------------------------------------
## treat: control
## vars n mean sd median trimmed mad min max range skew
## attitude 1 15 14.13 3.23 14 14.23 2.97 8 19 11 -0.31
## know 2 15 14.67 2.64 15 14.62 2.97 10 20 10 0.05
## kurtosis se
## attitude -0.99 0.83
## know -0.76 0.68Descriptive plots
- Scatterplot
library(ggplot2)
# The plot is colored by the group
scatterPlot <- ggplot(Hotellings_T, aes(attitude, know, color = treat)) +
geom_point() +
scale_color_manual(values = c('#999999','#E69F00')) +
labs(color = "treatment condition") +
theme_bw() +
theme(legend.position = "bottom")
scatterPlot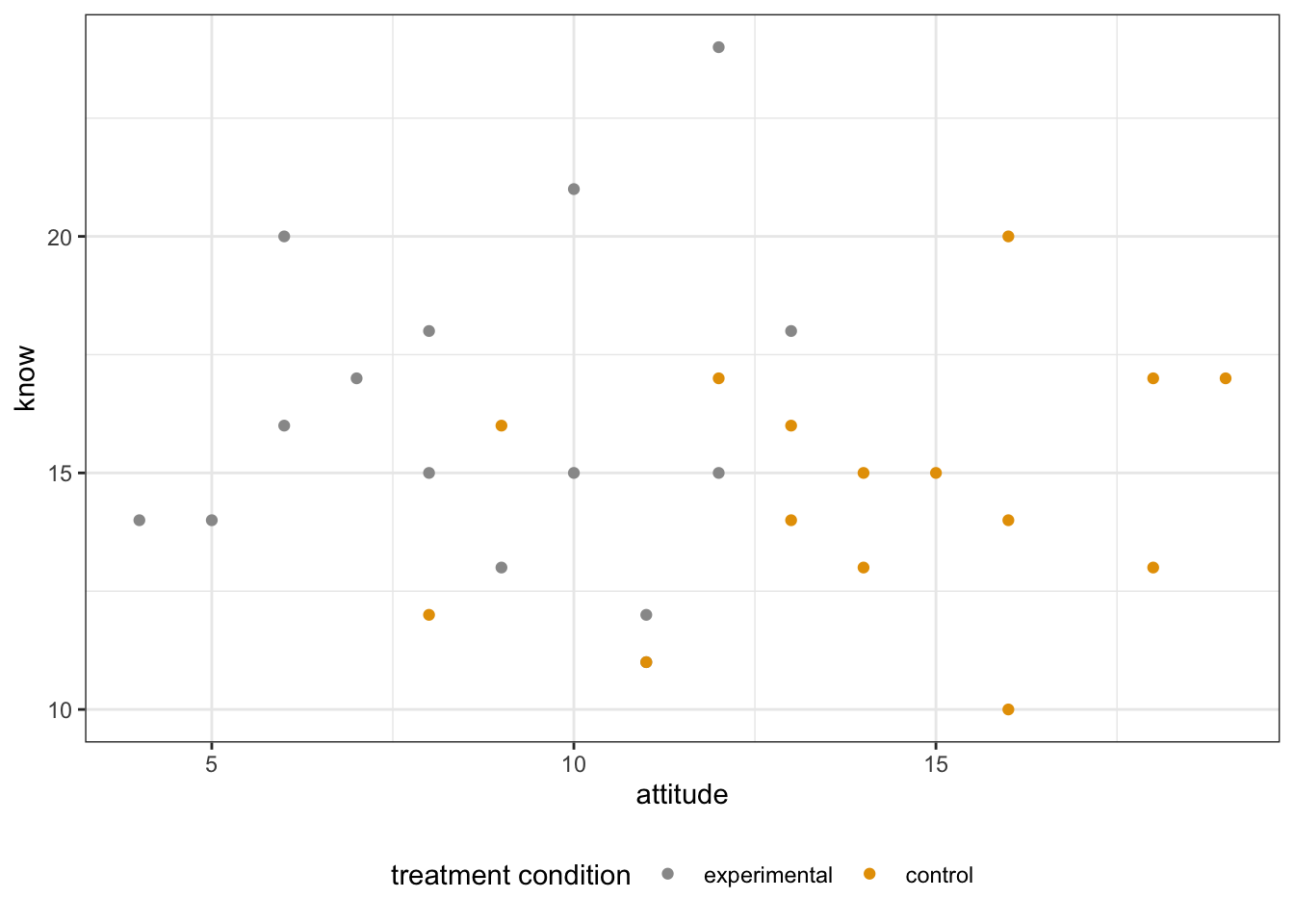
- Marginal density plot of attitude
xdensity <- ggplot(Hotellings_T, aes(attitude, fill=treat)) +
geom_density(alpha=.5) +
scale_fill_manual(values = c('#999999','#E69F00')) +
theme(legend.position = "none")
xdensity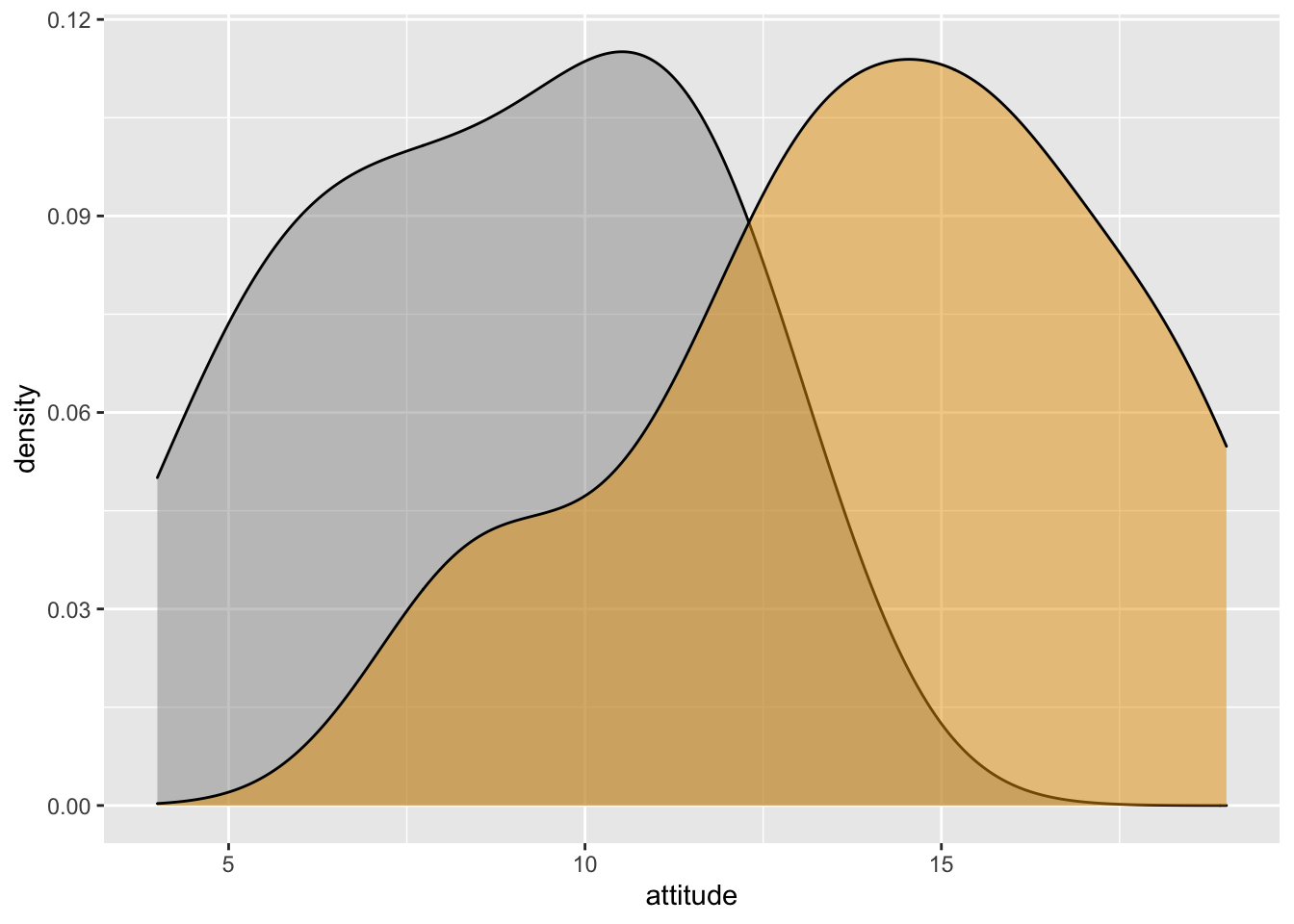
- Marginal density plot of knowledge
ydensity <- ggplot(Hotellings_T, aes(know, fill=treat)) +
geom_density(alpha=.5) +
scale_fill_manual(values = c('#999999','#E69F00')) +
theme(legend.position = "none")
ydensity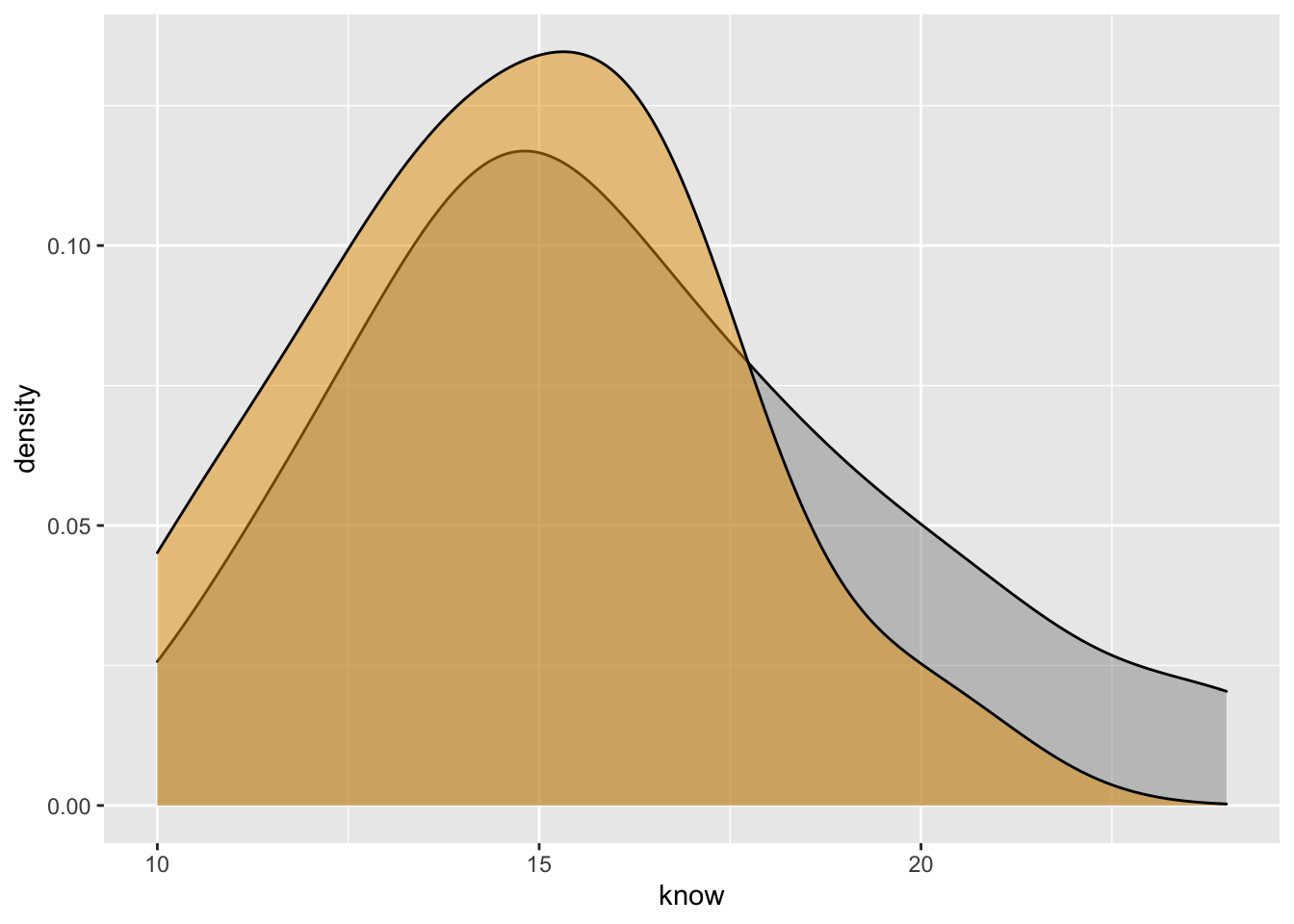
- Put them all together in one plot
blankPlot <- ggplot()+geom_blank(aes(1,1))+
theme(plot.background = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.border = element_blank(),
panel.background = element_blank(),
axis.title.x = element_blank(),
axis.title.y = element_blank(),
axis.text.x = element_blank(),
axis.text.y = element_blank(),
axis.ticks = element_blank()
)
library("gridExtra")
grid.arrange(xdensity, blankPlot, scatterPlot, ydensity,
ncol=2, nrow=2, widths=c(4, 1.4), heights=c(1.4, 4))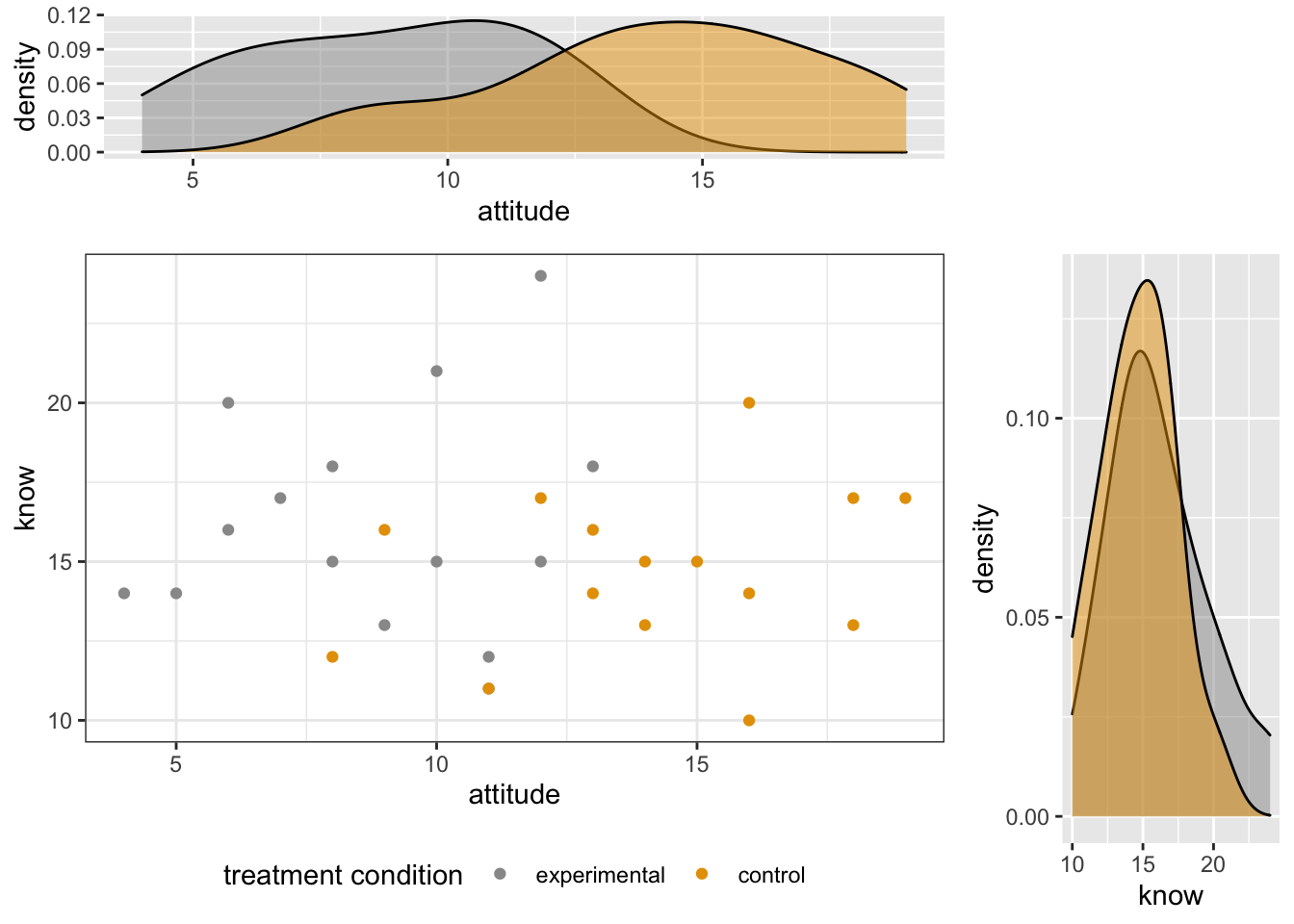
- You can also add centroids to the scatterplot:
centroids <- aggregate(cbind(attitude = attitude, know = know) ~ treat, data = Hotellings_T, mean)
scatterPlot + geom_point(centroids, mapping = aes(x = attitude, y = know), size = 5, shape = 17) +
labs(color = "treatment condition centroids")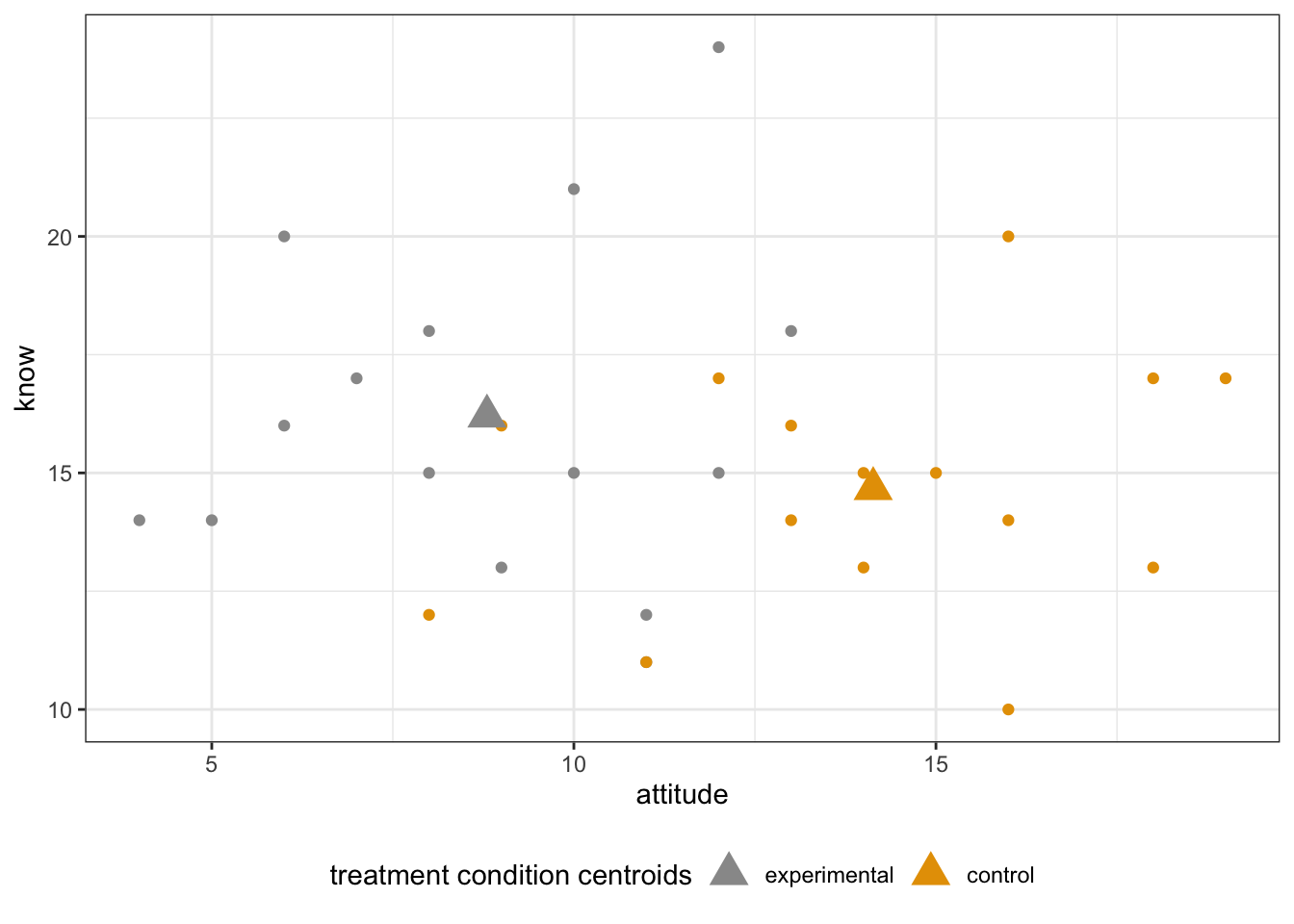
- You can also do boxplots in addition to density plots.
par(mfrow = c(2, 1), mar=c(1,1,1,1))
ggplot(Hotellings_T, aes(x=treat, y=attitude, color=treat)) +
geom_boxplot()+ theme(legend.position = "none") +
xlab("Treatment Condition")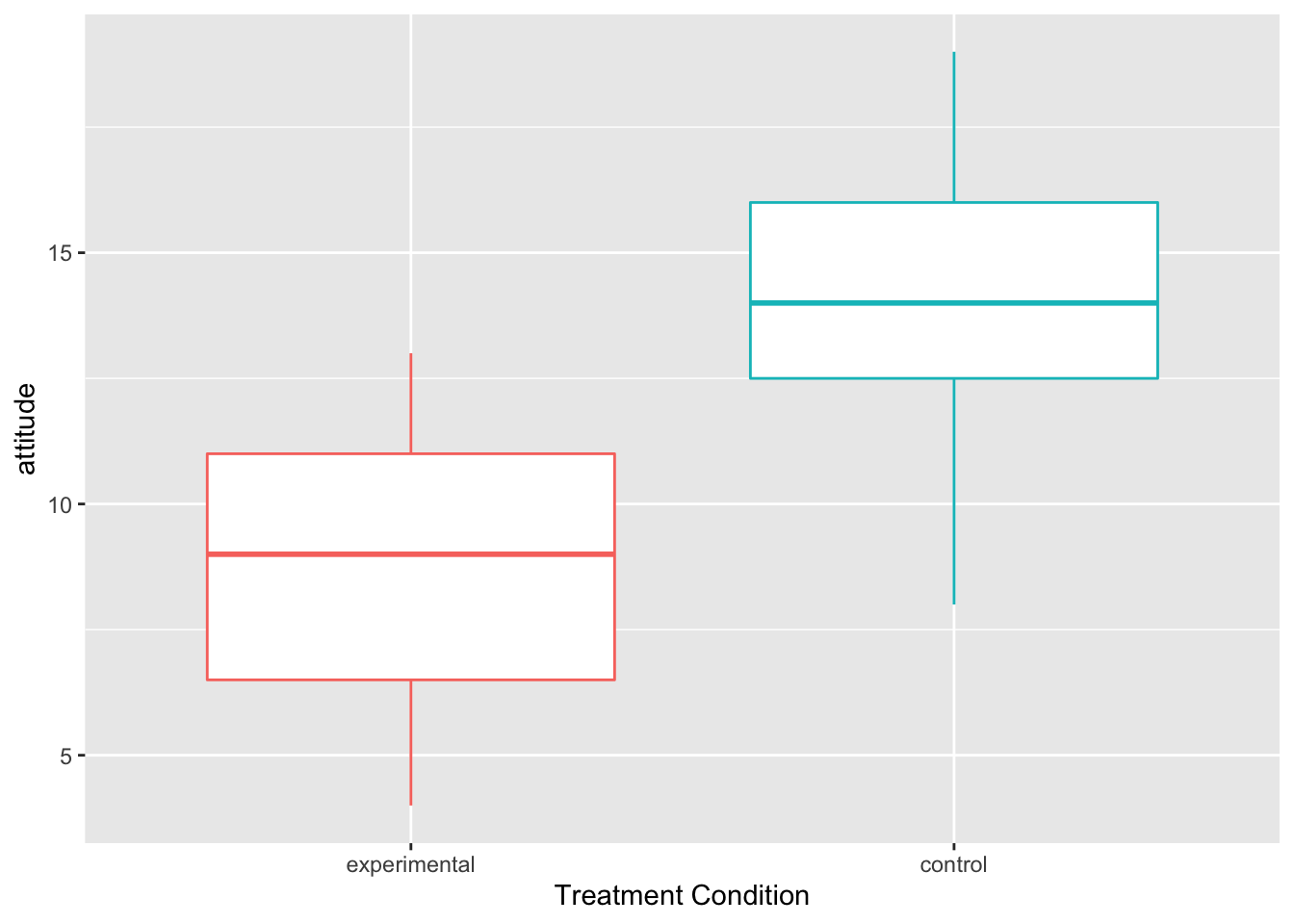
ggplot(Hotellings_T, aes(x=treat, y=know, color=treat)) +
geom_boxplot()+ theme(legend.position = "none") +
xlab("Treatment Condition")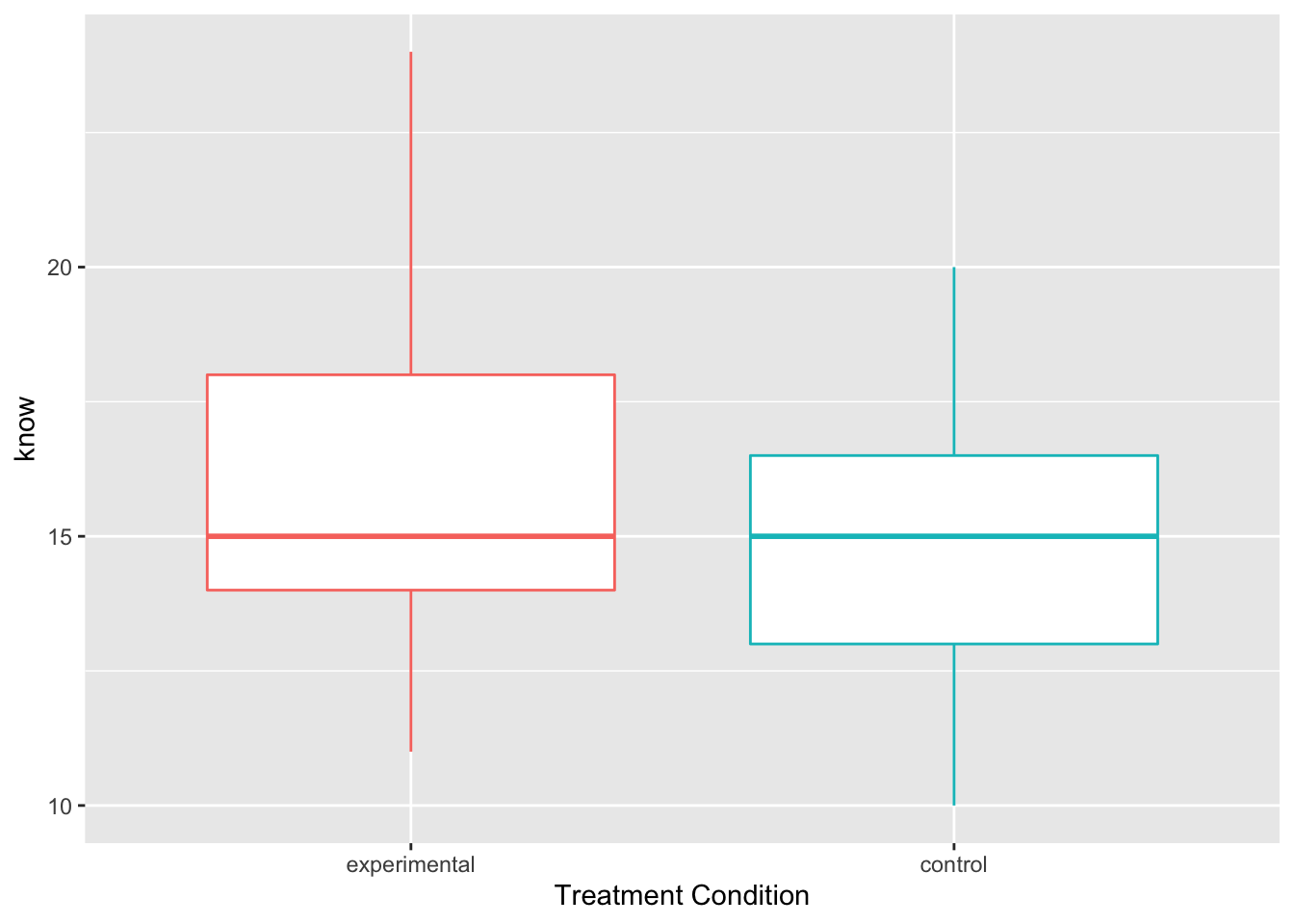
Step 3: Implement MANOVA analysis
Multivariate tests
You can obtain different types of statistics by specifying the
test= argument accordingly. The default will give you
Pillai’s trace.
manova.model1 <- manova(cbind(attitude = attitude, know = know) ~ treat, Hotellings_T)
summary(manova.model1, test = "Pillai", intercept=T)## Df Pillai approx F num Df den Df Pr(>F)
## (Intercept) 1 0.97268 480.57 2 27 < 2.2e-16 ***
## treat 1 0.50609 13.83 2 27 7.316e-05 ***
## Residuals 28
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(manova.model1, test = "Wilks", intercept=T)## Df Wilks approx F num Df den Df Pr(>F)
## (Intercept) 1 0.02732 480.57 2 27 < 2.2e-16 ***
## treat 1 0.49391 13.83 2 27 7.316e-05 ***
## Residuals 28
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(manova.model1, test = "Hotelling-Lawley", intercept=T)## Df Hotelling-Lawley approx F num Df den Df Pr(>F)
## (Intercept) 1 35.597 480.57 2 27 < 2.2e-16 ***
## treat 1 1.025 13.83 2 27 7.316e-05 ***
## Residuals 28
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(manova.model1, test = "Roy", intercept=T)## Df Roy approx F num Df den Df Pr(>F)
## (Intercept) 1 35.597 480.57 2 27 < 2.2e-16 ***
## treat 1 1.025 13.83 2 27 7.316e-05 ***
## Residuals 28
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1- Conduct the Box’s M test (homogeneity of dispersion)
library(heplots)
boxtest2 <- boxM(Hotellings_T[, c("attitude", "know")], Hotellings_T$treat)
summary(boxtest2)## Summary for Box's M-test of Equality of Covariance Matrices
##
## Chi-Sq: 1.544326
## df: 3
## p-value: 0.6721
##
## log of Covariance determinants:
## experimental control pooled
## 4.550595 4.217750 4.443953
##
## Eigenvalues:
## experimental control pooled
## 1 12.805776 11.4156 11.142702
## 2 7.394224 5.9463 7.638251
##
## Statistics based on eigenvalues:
## experimental control pooled
## product 94.688776 67.880612 85.110748
## sum 20.200000 17.361905 18.780952
## precision 4.687563 3.909745 4.531759
## max 12.805776 11.415604 11.142702Step 4: Follow-up: Univariate ANOVA
anova(lm(attitude~treat, Hotellings_T))## Analysis of Variance Table
##
## Response: attitude
## Df Sum Sq Mean Sq F value Pr(>F)
## treat 1 213.33 213.333 23.505 4.201e-05 ***
## Residuals 28 254.13 9.076
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1anova(lm(know~treat, Hotellings_T))## Analysis of Variance Table
##
## Response: know
## Df Sum Sq Mean Sq F value Pr(>F)
## treat 1 17.633 17.6333 1.817 0.1885
## Residuals 28 271.733 9.7048Example 2: Three-group MANOVA
Research Context
Middle school students are randomly assigned to one of three treatment groups. After receiving their respective treatment or control experience, students are evaluated in terms of cognitive and affective outcomes together. We are interested in testing whether there is any difference between the three treatment groups regarding the set of outcomes.
Step 1: Load the data
library(EDMS657Data)
data("manova_dat")take a look at the data you just read into R
#View(manova.data2)
glimpse(manova_dat)## Rows: 45
## Columns: 3
## $ treat <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2…
## $ attitude <dbl> 4, 5, 6, 6, 7, 8, 8, 9, 10, 10, 11, 11, 12, 12, 13, 8, …
## $ know <dbl> 14, 14, 16, 20, 17, 15, 18, 13, 15, 21, 11, 12, 15, 24,…str(manova_dat)## 'data.frame': 45 obs. of 3 variables:
## $ treat : num 1 1 1 1 1 1 1 1 1 1 ...
## ..- attr(*, "value.labels")= Named chr [1:3] "3" "2" "1"
## .. ..- attr(*, "names")= chr [1:3] "webinar" "control" "experimental"
## $ attitude: num 4 5 6 6 7 8 8 9 10 10 ...
## $ know : num 14 14 16 20 17 15 18 13 15 21 ...
## - attr(*, "variable.labels")= Named chr [1:3] "treatment condition" "attitude toward people with HIV/AIDS" "HIV/AIDS knowledge test"
## ..- attr(*, "names")= chr [1:3] "treat" "attitude" "know"
## - attr(*, "codepage")= int 65001head(manova_dat)## treat attitude know
## 1 1 4 14
## 2 1 5 14
## 3 1 6 16
## 4 1 6 20
## 5 1 7 17
## 6 1 8 15- Again, “treatment” is a categorical variable
manova_dat$treat <- as.factor(manova_dat$treat)
levels(manova_dat$treat) <- c("experimental", "control", "webinar")Step 2: Explore the data with descriptive statistics and plots
Descriptive statistics
summary(manova_dat)## treat attitude know
## experimental:15 Min. : 4.000 Min. : 8.00
## control :15 1st Qu.: 7.000 1st Qu.:11.00
## webinar :15 Median : 9.000 Median :14.00
## Mean : 9.933 Mean :14.02
## 3rd Qu.:13.000 3rd Qu.:16.00
## Max. :19.000 Max. :24.00table(manova_dat$treat)##
## experimental control webinar
## 15 15 15Within-group means
with(manova_dat, tapply(attitude, treat, mean))## experimental control webinar
## 8.800000 14.133333 6.866667with(manova_dat, tapply(know, treat, mean))## experimental control webinar
## 16.20000 14.66667 11.20000Within-group variances
with(manova_dat, tapply(attitude, treat, var))## experimental control webinar
## 7.742857 10.409524 4.552381with(manova_dat, tapply(know, treat, var))## experimental control webinar
## 12.457143 6.952381 3.314286psych::describeBy(attitude + know ~ treat, data = manova_dat)##
## Descriptive statistics by group
## treat: experimental
## vars n mean sd median trimmed mad min max range skew
## attitude 1 15 8.8 2.78 9 8.85 2.97 4 13 9 -0.16
## know 2 15 16.2 3.53 15 16.00 2.97 11 24 13 0.57
## kurtosis se
## attitude -1.39 0.72
## know -0.57 0.91
## -------------------------------------------------------
## treat: control
## vars n mean sd median trimmed mad min max range skew
## attitude 1 15 14.13 3.23 14 14.23 2.97 8 19 11 -0.31
## know 2 15 14.67 2.64 15 14.62 2.97 10 20 10 0.05
## kurtosis se
## attitude -0.99 0.83
## know -0.76 0.68
## -------------------------------------------------------
## treat: webinar
## vars n mean sd median trimmed mad min max range skew
## attitude 1 15 6.87 2.13 7 6.77 2.97 4 11 7 0.33
## know 2 15 11.20 1.82 11 11.23 1.48 8 14 6 -0.01
## kurtosis se
## attitude -1.11 0.55
## know -1.18 0.47Descriptive plots
Scatterplot
library(ggplot2)
scatterPlot2 <- ggplot(manova_dat, aes(attitude, know, color=treat)) +
geom_point() +
scale_color_manual(values = c("#999999", "#E69F00", "#56B4E9")) +
labs(color="treatment condition") +
theme_bw() +
theme(legend.position = "bottom")
scatterPlot2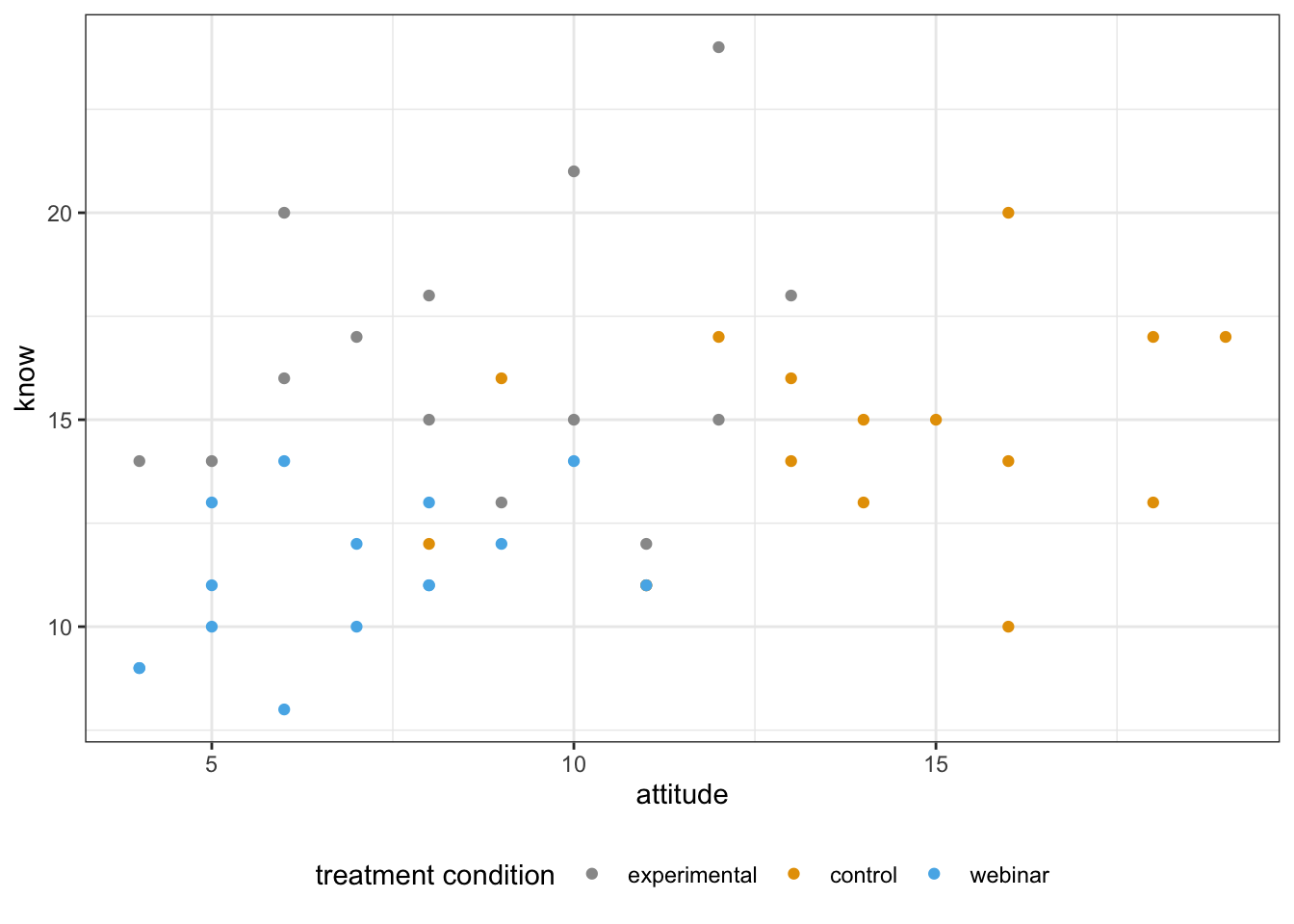
xdensity2 <- ggplot(manova_dat, aes(attitude, fill=treat)) +
geom_density(alpha=.5) +
scale_fill_manual(values = c("#999999", "#E69F00", "#56B4E9")) +
theme(legend.position = "none")
xdensity2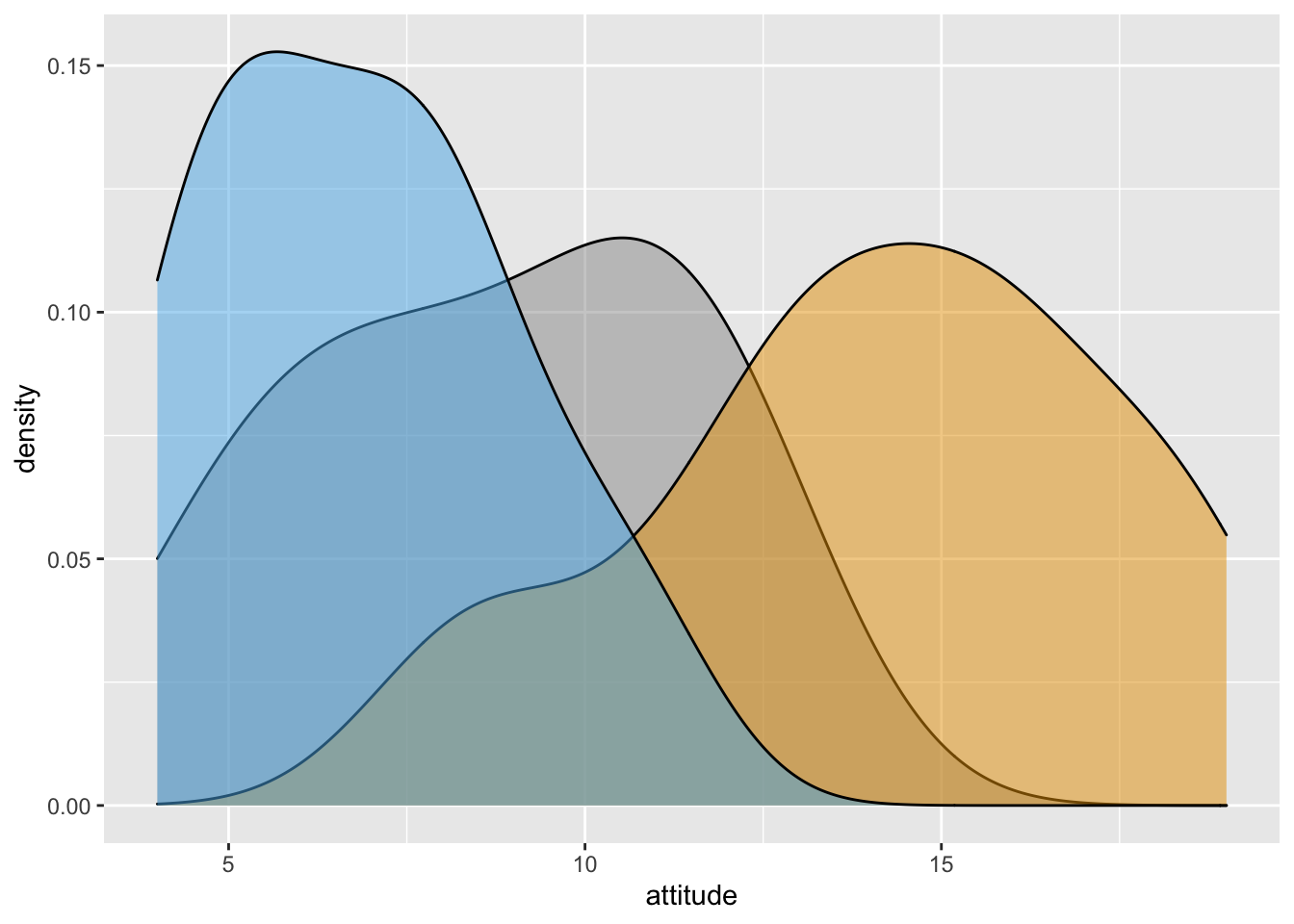
ydensity2 <- ggplot(manova_dat, aes(know, fill=treat)) +
geom_density(alpha=.5) +
scale_fill_manual(values = c("#999999", "#E69F00", "#56B4E9")) +
theme(legend.position = "none")
ydensity2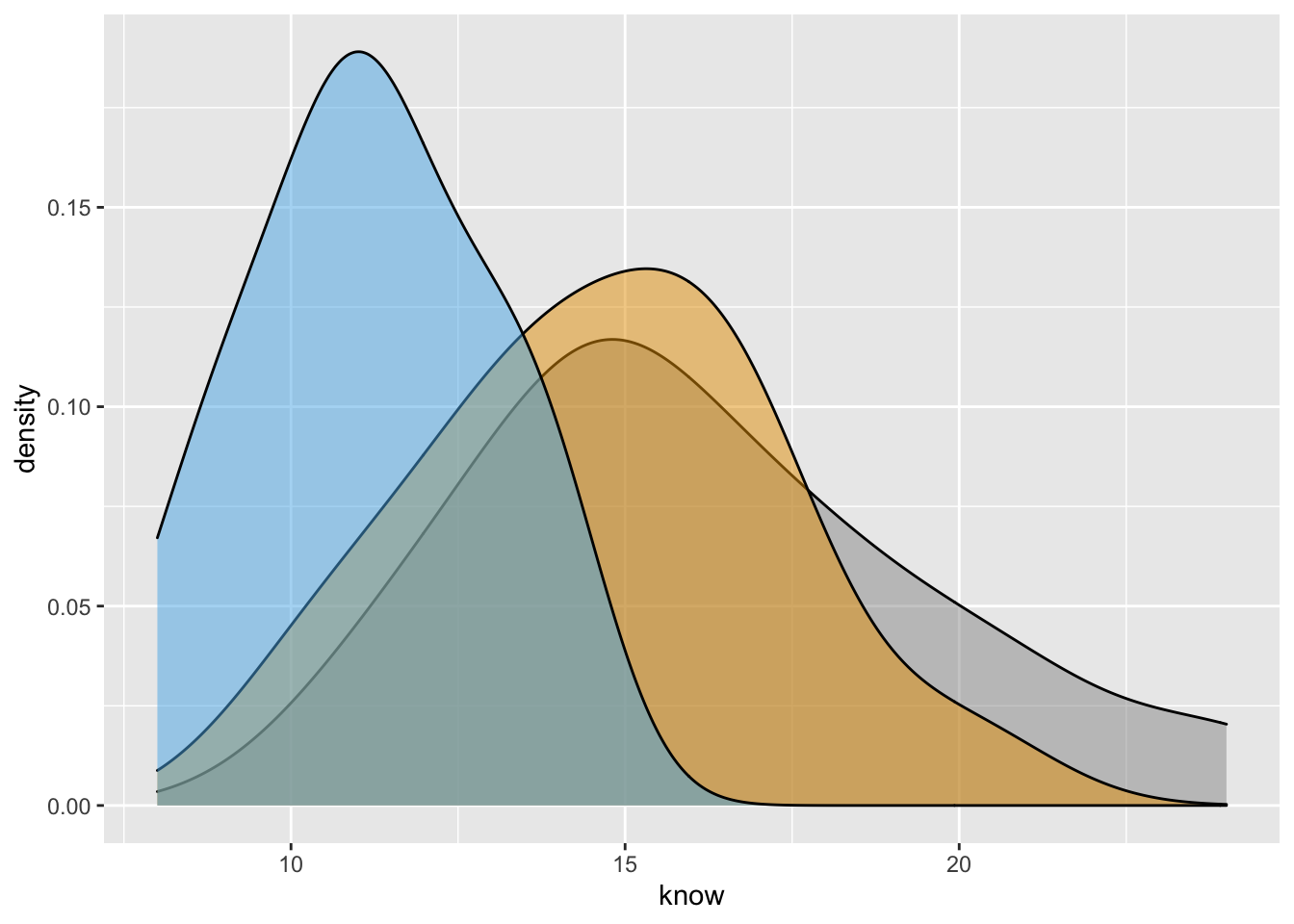
blankPlot <- ggplot()+geom_blank(aes(1,1))+
theme(plot.background = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.border = element_blank(),
panel.background = element_blank(),
axis.title.x = element_blank(),
axis.title.y = element_blank(),
axis.text.x = element_blank(),
axis.text.y = element_blank(),
axis.ticks = element_blank()
)
library("gridExtra")
grid.arrange(xdensity2, blankPlot, scatterPlot2, ydensity2,
ncol=2, nrow=2, widths=c(4, 1.4), heights=c(1.4, 4))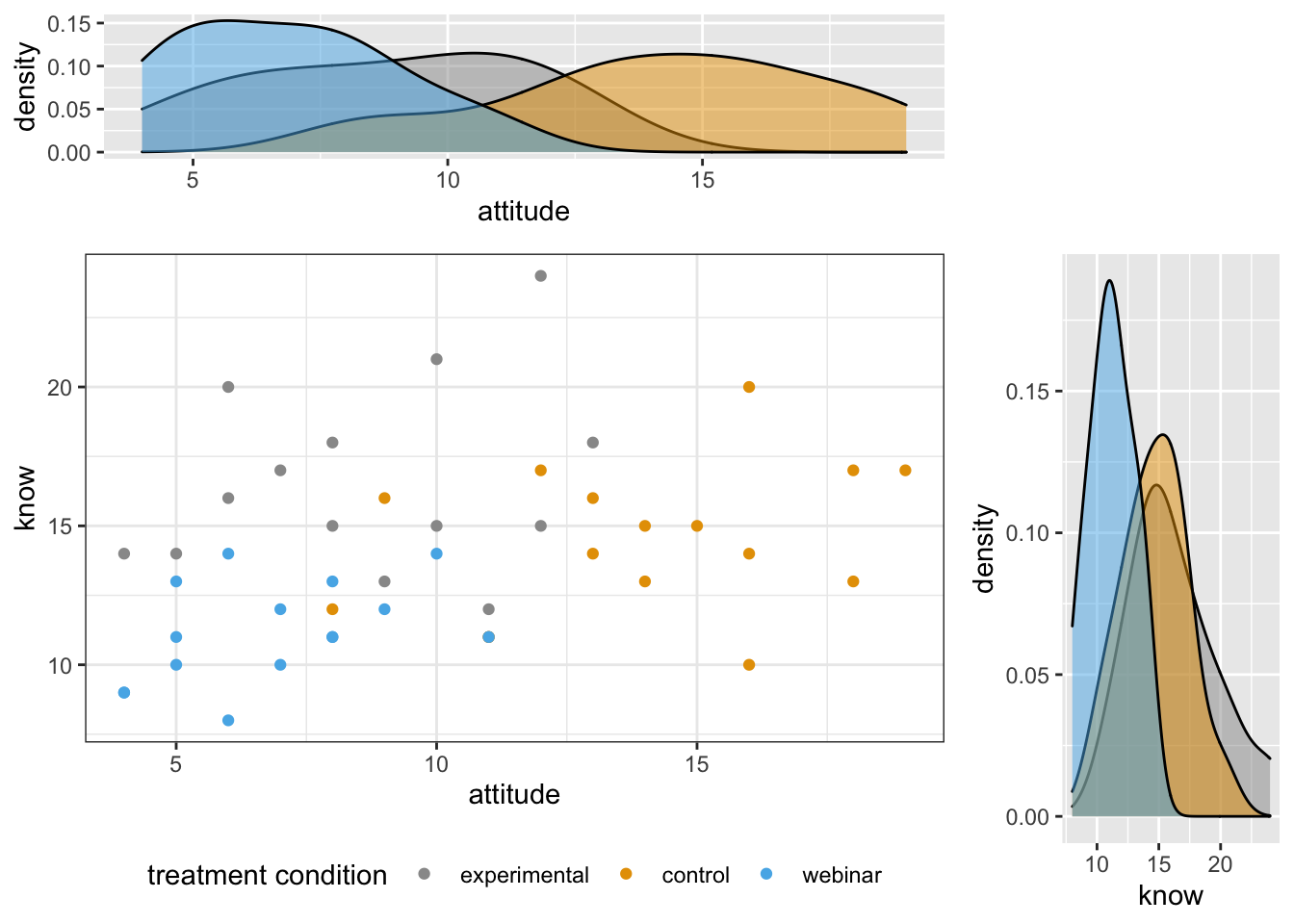
Add centroids
centroids2 <- aggregate(cbind(attitude = attitude, know = know)~treat, data = manova_dat, mean)
scatterPlot2 + geom_point(centroids2,mapping=aes(x=attitude,y=know),size=5,shape=17)+
labs(color="treatment condition centroids")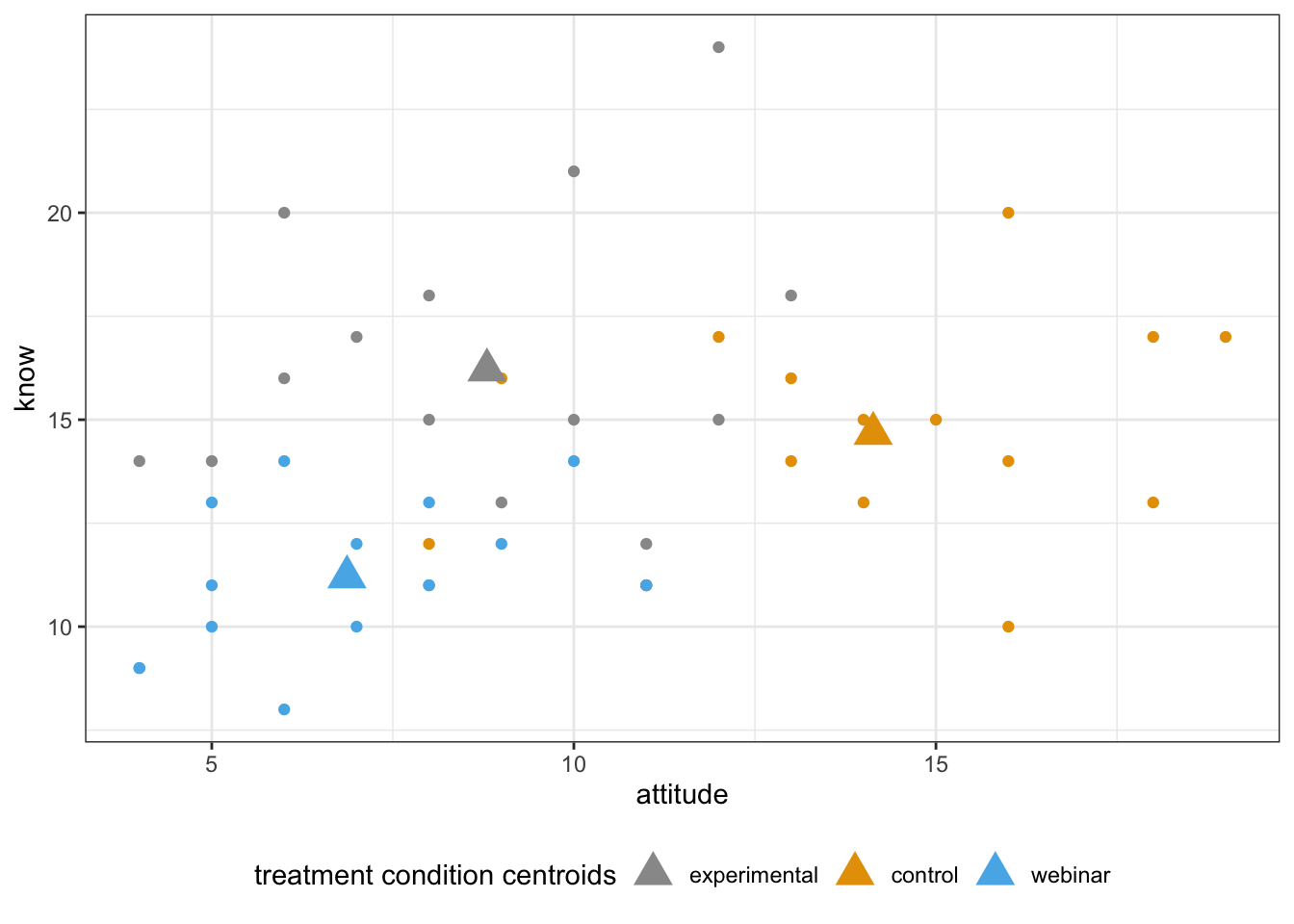
Boxplots
ggplot(manova_dat, aes(x=treat, y=attitude, color=treat)) +
geom_boxplot()+ theme(legend.position = "none") +
xlab("Treatment Condition")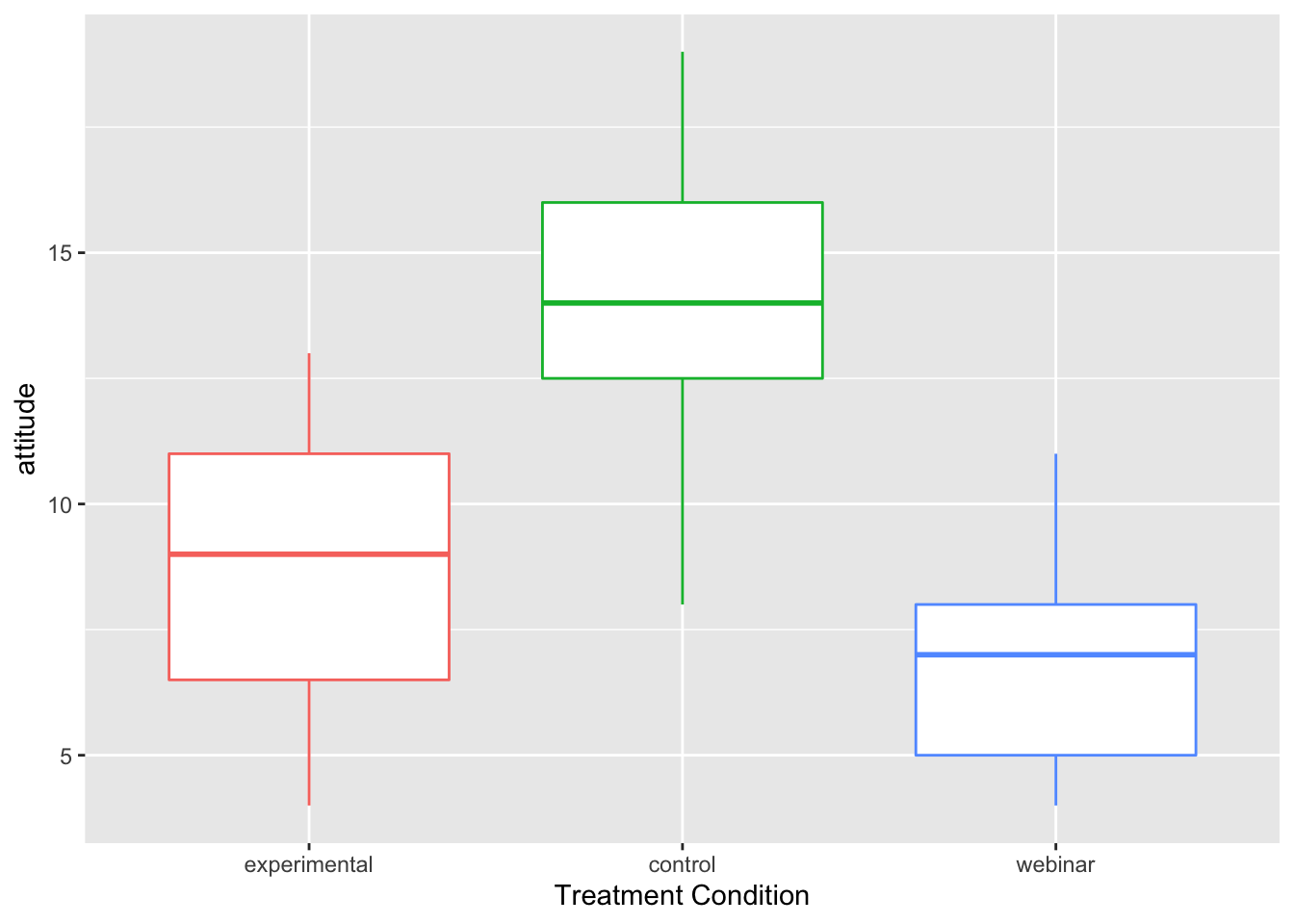
ggplot(manova_dat, aes(x=treat, y=know, color=treat)) +
geom_boxplot()+ theme(legend.position = "none") +
xlab("Treatment Condition")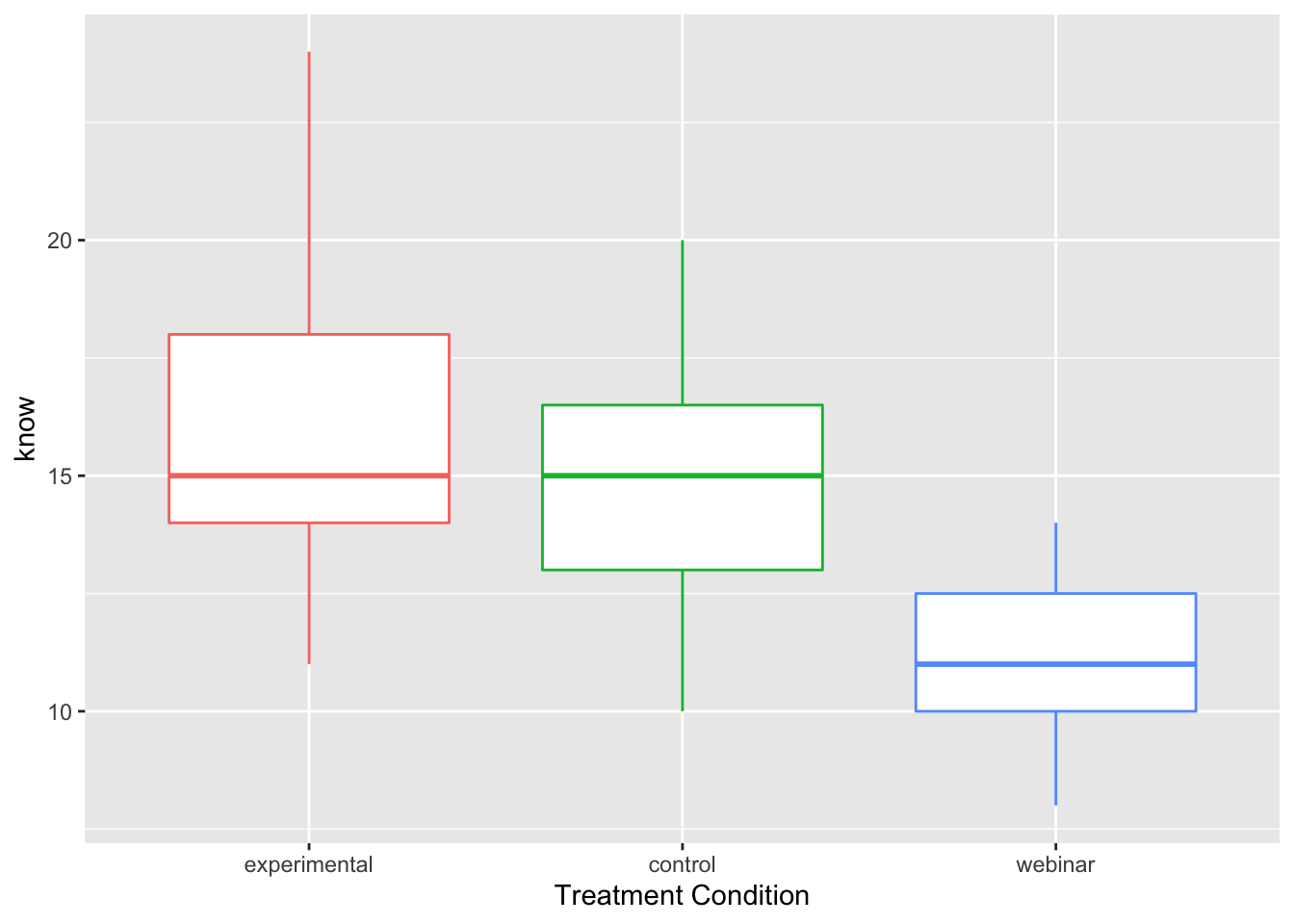
Raincloud plots
Inspired by this online post by Dr. Allen, we could do raincloud plots to describe the data.
source("https://gist.githubusercontent.com/benmarwick/2a1bb0133ff568cbe28d/raw/fb53bd97121f7f9ce947837ef1a4c65a73bffb3f/geom_flat_violin.R")
raincloud_theme = theme(
text = element_text(size = 10),
axis.title.x = element_text(size = 16),
axis.title.y = element_text(size = 16),
axis.text = element_text(size = 14),
axis.text.x = element_text(angle = 45, vjust = 0.5),
legend.title=element_text(size=16),
legend.text=element_text(size=16),
legend.position = "right",
plot.title = element_text(lineheight=.8, face="bold", size = 16),
panel.border = element_blank(),
panel.grid.minor = element_blank(),
panel.grid.major = element_blank(),
axis.line.x = element_line(colour = 'black', size=0.5, linetype='solid'),
axis.line.y = element_line(colour = 'black', size=0.5, linetype='solid'))
rc_plot <- ggplot(data = manova_dat, aes(x=treat, y=attitude, fill = treat)) +
geom_flat_violin(position = position_nudge(x = .2, y = 0), alpha = .8) +
geom_point(aes(y=attitude, color = treat), position = position_jitter(width = .15), size = .5, alpha = 0.8) +
geom_boxplot(width = .1, guides = FALSE, outlier.shape = NA, alpha = 0.5) +
expand_limits(x = 4, y = 20) +
guides(fill = FALSE) +
guides(color = FALSE) +
scale_color_brewer(palette = "Set1") +
scale_fill_brewer(palette = "Set1") +
theme_bw() +
raincloud_theme
rc_plot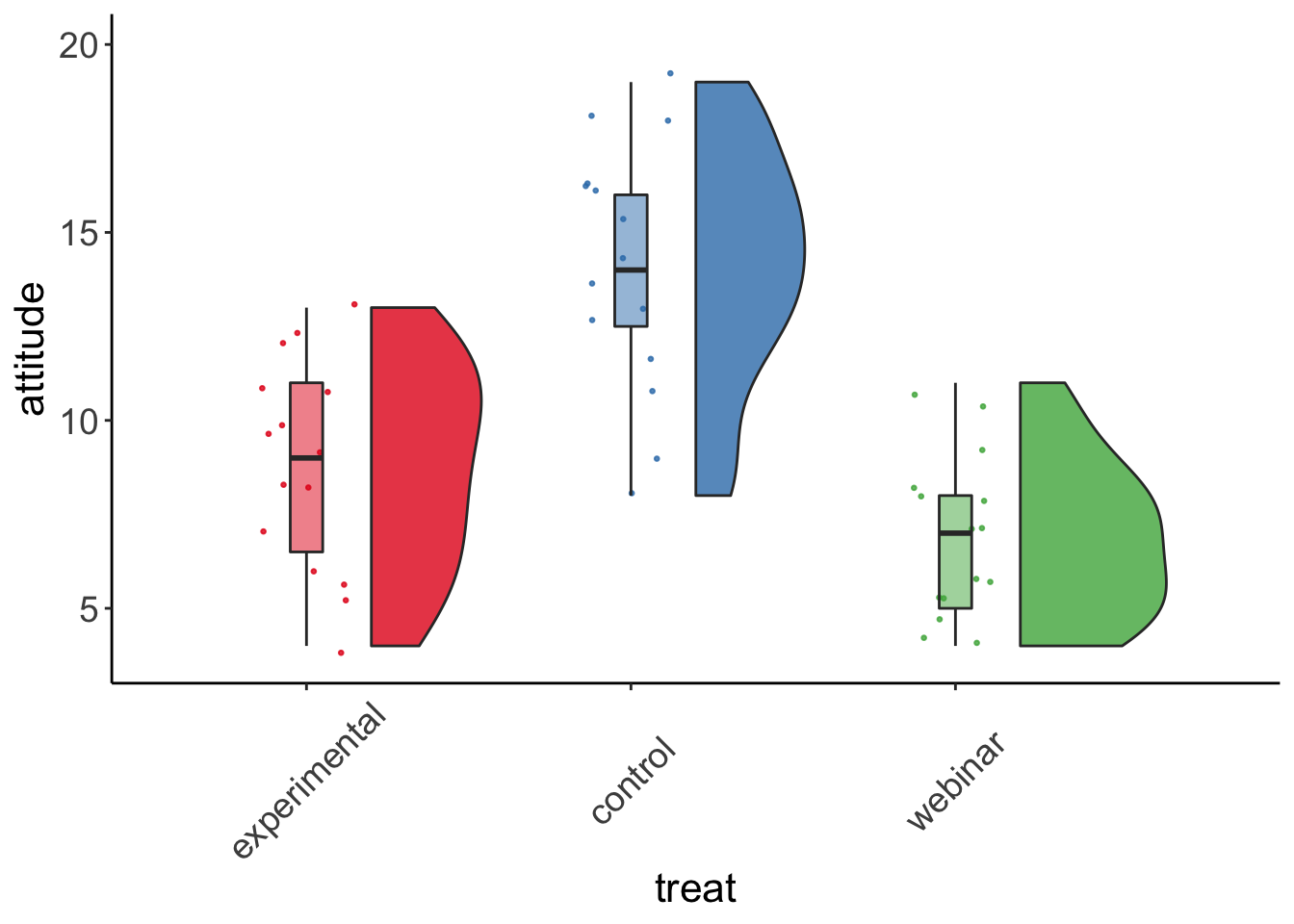
rc_plot2 <- ggplot(data = manova_dat, aes(x=treat, y=know, fill = treat)) +
geom_flat_violin(position = position_nudge(x = .2, y = 0), alpha = .8) +
geom_point(aes(y=know, color = treat), position = position_jitter(width = .15), size = .5, alpha = 0.8) +
geom_boxplot(width = .1, guides = FALSE, outlier.shape = NA, alpha = 0.5) +
expand_limits(x = 4, y = 20) +
guides(fill = FALSE) +
guides(color = FALSE) +
scale_color_brewer(palette = "Set1") +
scale_fill_brewer(palette = "Set1") +
theme_bw() +
raincloud_theme
rc_plot2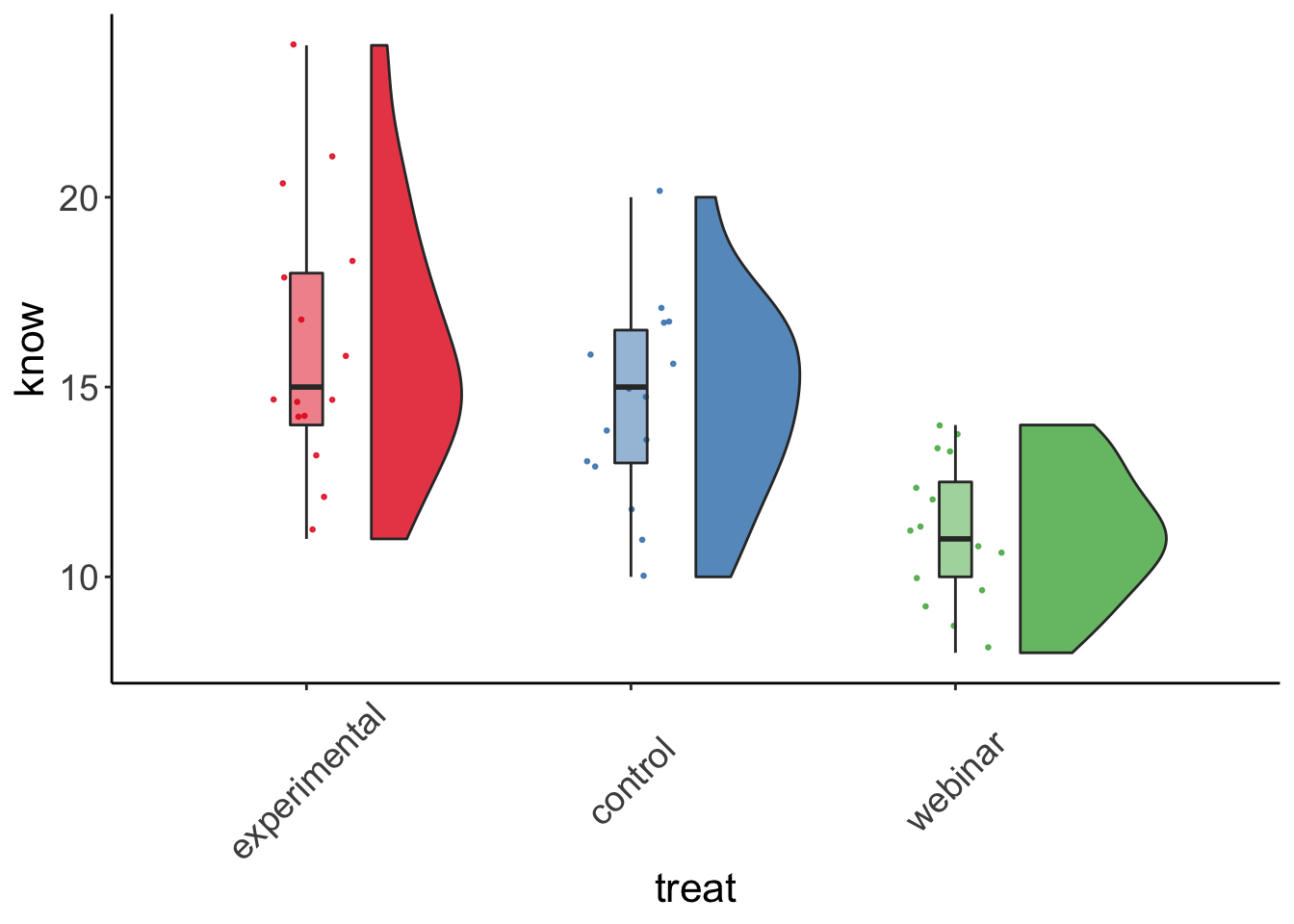
Step 3: Implement MANOVA
Multivariate tests
manova.model2 <- manova(cbind(attitude = attitude, know = know) ~ treat, manova_dat)
summary(manova.model2, test = "Pillai", intercept=T)## Df Pillai approx F num Df den Df Pr(>F)
## (Intercept) 1 0.97196 710.47 2 41 < 2.2e-16 ***
## treat 2 0.91148 17.58 4 84 1.614e-10 ***
## Residuals 42
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(manova.model2, test = "Wilks", intercept=T)## Df Wilks approx F num Df den Df Pr(>F)
## (Intercept) 1 0.028045 710.47 2 41 < 2.2e-16 ***
## treat 2 0.281925 18.11 4 82 1.081e-10 ***
## Residuals 42
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(manova.model2, test = "Hotelling-Lawley", intercept=T)## Df Hotelling-Lawley approx F num Df den Df Pr(>F)
## (Intercept) 1 34.657 710.47 2 41 < 2.2e-16 ***
## treat 2 1.861 18.61 4 80 7.589e-11 ***
## Residuals 42
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(manova.model2, test = "Roy", intercept=T)## Df Roy approx F num Df den Df Pr(>F)
## (Intercept) 1 34.657 710.47 2 41 < 2.2e-16 ***
## treat 2 1.355 28.45 2 42 1.548e-08 ***
## Residuals 42
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Box’s M test
library(heplots)
boxtest4 <- boxM(manova_dat[,c("attitude","know")], manova_dat$treat)
summary(boxtest4)## Summary for Box's M-test of Equality of Covariance Matrices
##
## Chi-Sq: 9.288089
## df: 6
## p-value: 0.158
##
## log of Covariance determinants:
## experimental control webinar pooled
## 4.550595 4.217750 2.509129 3.996638
##
## Eigenvalues:
## experimental control webinar pooled
## 1 12.805776 11.4156 5.715718 9.277781
## 2 7.394224 5.9463 2.150949 5.865076
##
## Statistics based on eigenvalues:
## experimental control webinar pooled
## product 94.688776 67.880612 12.294218 54.414893
## sum 20.200000 17.361905 7.866667 15.142857
## precision 4.687563 3.909745 1.562824 3.593436
## max 12.805776 11.415604 5.715718 9.277781Step 4: Follow-up analyses
Univariate ANOVA
aov.att <- aov(attitude~treat, manova_dat)
summary(aov.att)## Df Sum Sq Mean Sq F value Pr(>F)
## treat 2 424.9 212.47 28.07 1.81e-08 ***
## Residuals 42 317.9 7.57
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1aov.kn <- aov(know~treat, manova_dat)
summary(aov.kn)## Df Sum Sq Mean Sq F value Pr(>F)
## treat 2 196.8 98.42 12.99 4.05e-05 ***
## Residuals 42 318.1 7.57
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1LSD test
Because the test results for the univariate ANOVA are statistically significant, we can proceed to conducting LSD test for multiple comparisons.
library(agricolae)
lsd.test.att <- LSD.test(aov.att, "treat")
lsd.test.kn <- LSD.test(aov.kn, "treat")
lsd.test.att$groups## attitude groups
## control 14.133333 a
## experimental 8.800000 b
## webinar 6.866667 blsd.test.kn$groups## know groups
## experimental 16.20000 a
## control 14.66667 a
## webinar 11.20000 b“Manual” multivariate multiple comparisons
summary(manova(cbind(attitude = attitude, know = know) ~ treat, manova_dat[manova_dat$treat%in%c("experimental","control"),]), test = "Wilks", intercept = T)## Df Wilks approx F num Df den Df Pr(>F)
## (Intercept) 1 0.02732 480.57 2 27 < 2.2e-16 ***
## treat 1 0.49391 13.83 2 27 7.316e-05 ***
## Residuals 28
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(manova(cbind(attitude = attitude, know = know) ~ treat, manova_dat[manova_dat$treat%in%c("experimental","webinar"),]), test = "Wilks", intercept = T)## Df Wilks approx F num Df den Df Pr(>F)
## (Intercept) 1 0.03175 411.69 2 27 < 2.2e-16 ***
## treat 1 0.52819 12.06 2 27 0.000181 ***
## Residuals 28
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(manova(cbind(attitude = attitude, know = know) ~ treat, manova_dat[manova_dat$treat%in%c("webinar","control"),]), test = "Wilks", intercept = T)## Df Wilks approx F num Df den Df Pr(>F)
## (Intercept) 1 0.02434 541.23 2 27 < 2.2e-16 ***
## treat 1 0.32864 27.58 2 27 2.991e-07 ***
## Residuals 28
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Example 3: Three-group MANOVA with six outcomes
Research Context
The faculties from a public research univeristy were evaluated at the end of a semester with regard to the courses they taught in that semester. Students rated the faculties on six aspects. We are interested in testing whether there is any difference between different types of faculties regarding the set of outcomes.
Step 1: Load the data
library(EDMS657Data)
data(Teacher_ratings)take a look at the data you just read into R
#View(Teacher_ratings)
glimpse(Teacher_ratings)## Rows: 120
## Columns: 7
## $ organize <dbl> 4, 7, 6, 6, 4, 8, 2, 4, 4, 6, 7, 7, 4, 5, 5, 5, 3, 6, 7…
## $ clarity <dbl> 4, 7, 6, 8, 3, 6, 3, 2, 5, 6, 7, 8, 5, 5, 5, 3, 4, 7, 7…
## $ interact <dbl> 3, 6, 5, 6, 5, 8, 3, 2, 6, 6, 4, 6, 4, 6, 4, 5, 5, 5, 7…
## $ workload <dbl> 6, 4, 4, 6, 4, 6, 3, 2, 6, 5, 4, 5, 6, 3, 4, 7, 5, 3, 4…
## $ grading <dbl> 4, 4, 5, 6, 4, 6, 5, 2, 5, 5, 3, 6, 5, 2, 4, 6, 3, 3, 3…
## $ learning <dbl> 3, 4, 6, 5, 1, 7, 2, 3, 4, 4, 4, 5, 6, 2, 5, 6, 3, 2, 4…
## $ group <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1…str(Teacher_ratings)## 'data.frame': 120 obs. of 7 variables:
## $ organize: num 4 7 6 6 4 8 2 4 4 6 ...
## $ clarity : num 4 7 6 8 3 6 3 2 5 6 ...
## $ interact: num 3 6 5 6 5 8 3 2 6 6 ...
## $ workload: num 6 4 4 6 4 6 3 2 6 5 ...
## $ grading : num 4 4 5 6 4 6 5 2 5 5 ...
## $ learning: num 3 4 6 5 1 7 2 3 4 4 ...
## $ group : num 1 1 1 1 1 1 1 1 1 1 ...
## ..- attr(*, "value.labels")= Named chr [1:3] "3" "2" "1"
## .. ..- attr(*, "names")= chr [1:3] "Full" "Associate" "Assistant"
## - attr(*, "variable.labels")= Named chr [1:7] "course organization and planning" "clarity and communication skills" "teacher/student interaction and rapport" "course difficulty and workload" ...
## ..- attr(*, "names")= chr [1:7] "organize" "clarity" "interact" "workload" ...
## - attr(*, "codepage")= int 65001head(Teacher_ratings)## organize clarity interact workload grading learning group
## 1 4 4 3 6 4 3 1
## 2 7 7 6 4 4 4 1
## 3 6 6 5 4 5 6 1
## 4 6 8 6 6 6 5 1
## 5 4 3 5 4 4 1 1
## 6 8 6 8 6 6 7 1“treatment” is a categorical variable
Teacher_ratings$group <- as.factor(Teacher_ratings$group)
levels(Teacher_ratings$group) <- c("assistant", "associate", "full")Step 2: Explore the data with descriptive statistics and plots
Descriptive statistics
summary(Teacher_ratings)## organize clarity interact workload
## Min. :1.0 Min. :1.000 Min. :1.000 Min. :1.00
## 1st Qu.:4.0 1st Qu.:4.000 1st Qu.:4.000 1st Qu.:3.00
## Median :5.0 Median :5.000 Median :5.000 Median :4.00
## Mean :5.1 Mean :5.142 Mean :4.975 Mean :4.35
## 3rd Qu.:6.0 3rd Qu.:6.000 3rd Qu.:6.000 3rd Qu.:5.00
## Max. :8.0 Max. :9.000 Max. :9.000 Max. :8.00
## grading learning group
## Min. :1.000 Min. :1.00 assistant:40
## 1st Qu.:3.000 1st Qu.:3.00 associate:40
## Median :4.000 Median :4.00 full :40
## Mean :4.225 Mean :4.45
## 3rd Qu.:5.000 3rd Qu.:5.00
## Max. :9.000 Max. :9.00table(Teacher_ratings$group)##
## assistant associate full
## 40 40 40Within-group means
for (i in 1:6){
print(colnames(Teacher_ratings)[i])
print(with(Teacher_ratings, tapply(Teacher_ratings[,i], group, mean)))
}## [1] "organize"
## assistant associate full
## 5.075 5.725 4.500
## [1] "clarity"
## assistant associate full
## 5.200 5.875 4.350
## [1] "interact"
## assistant associate full
## 4.875 5.750 4.300
## [1] "workload"
## assistant associate full
## 4.40 5.00 3.65
## [1] "grading"
## assistant associate full
## 4.125 5.025 3.525
## [1] "learning"
## assistant associate full
## 4.275 5.250 3.825Within-group variances
for (i in 1:6){
print(colnames(Teacher_ratings)[i])
print(with(Teacher_ratings, tapply(Teacher_ratings[,i], group, var)))
}## [1] "organize"
## assistant associate full
## 3.096795 1.896795 1.948718
## [1] "clarity"
## assistant associate full
## 2.882051 1.958333 2.130769
## [1] "interact"
## assistant associate full
## 2.727564 2.397436 2.830769
## [1] "workload"
## assistant associate full
## 2.451282 3.333333 1.156410
## [1] "grading"
## assistant associate full
## 1.548077 2.486538 1.025000
## [1] "learning"
## assistant associate full
## 2.2044872 2.5000000 0.9685897Descriptive plots
Let’s try something different from the previous two examples. The following plot shows the covariance ellipse for each group.
library(heplots)
covEllipses(Teacher_ratings[,1:6], Teacher_ratings$group,
fill=c(rep(FALSE,3), TRUE), pooled=F)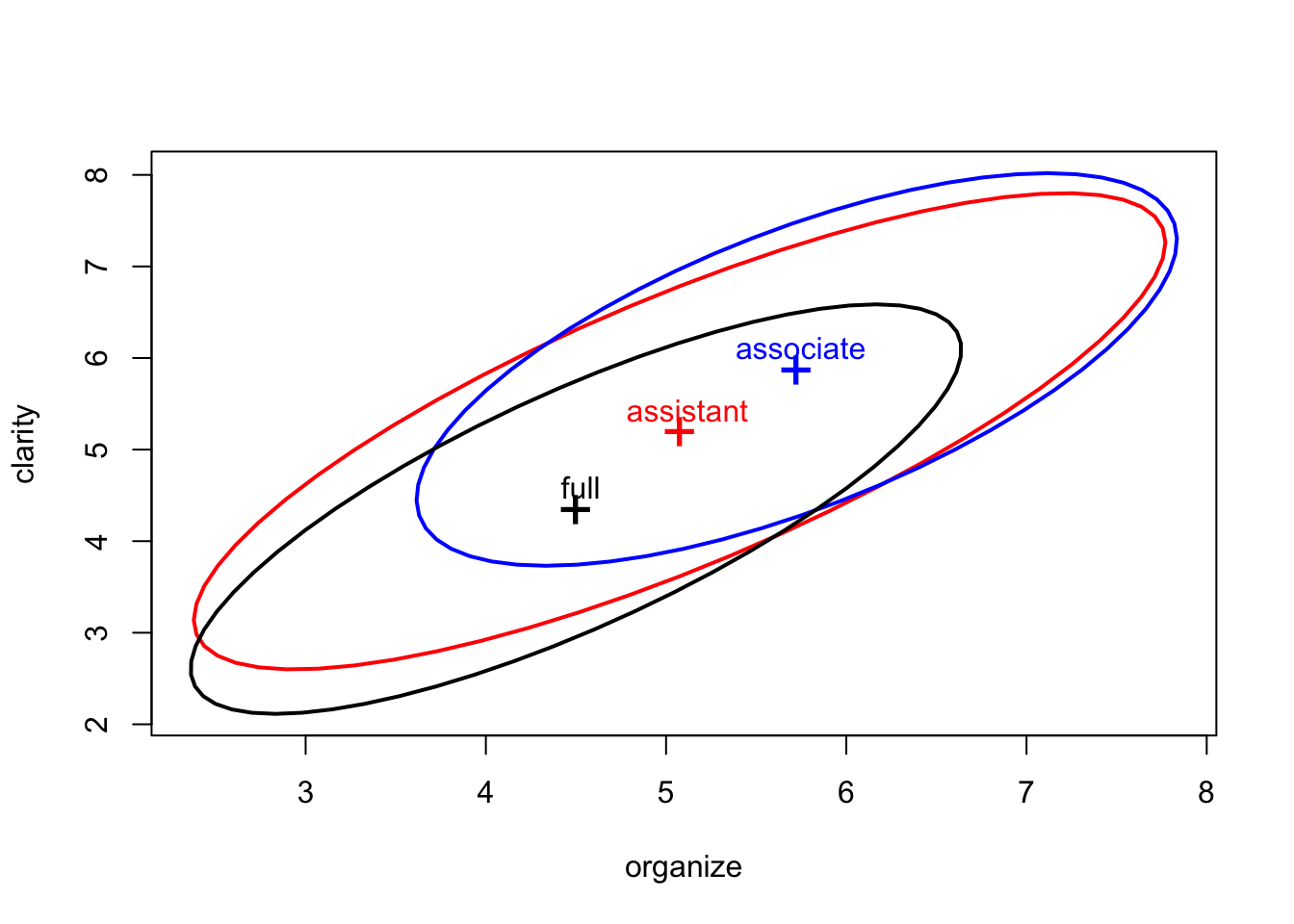
You can center the covariance ellipses by setting a common centroid for all the groups.
covEllipses(Teacher_ratings[,1:6], Teacher_ratings$group, center=TRUE, fill=c(rep(FALSE,3), TRUE), fill.alpha=.1, label.pos=c(1:3,0), pooled=F)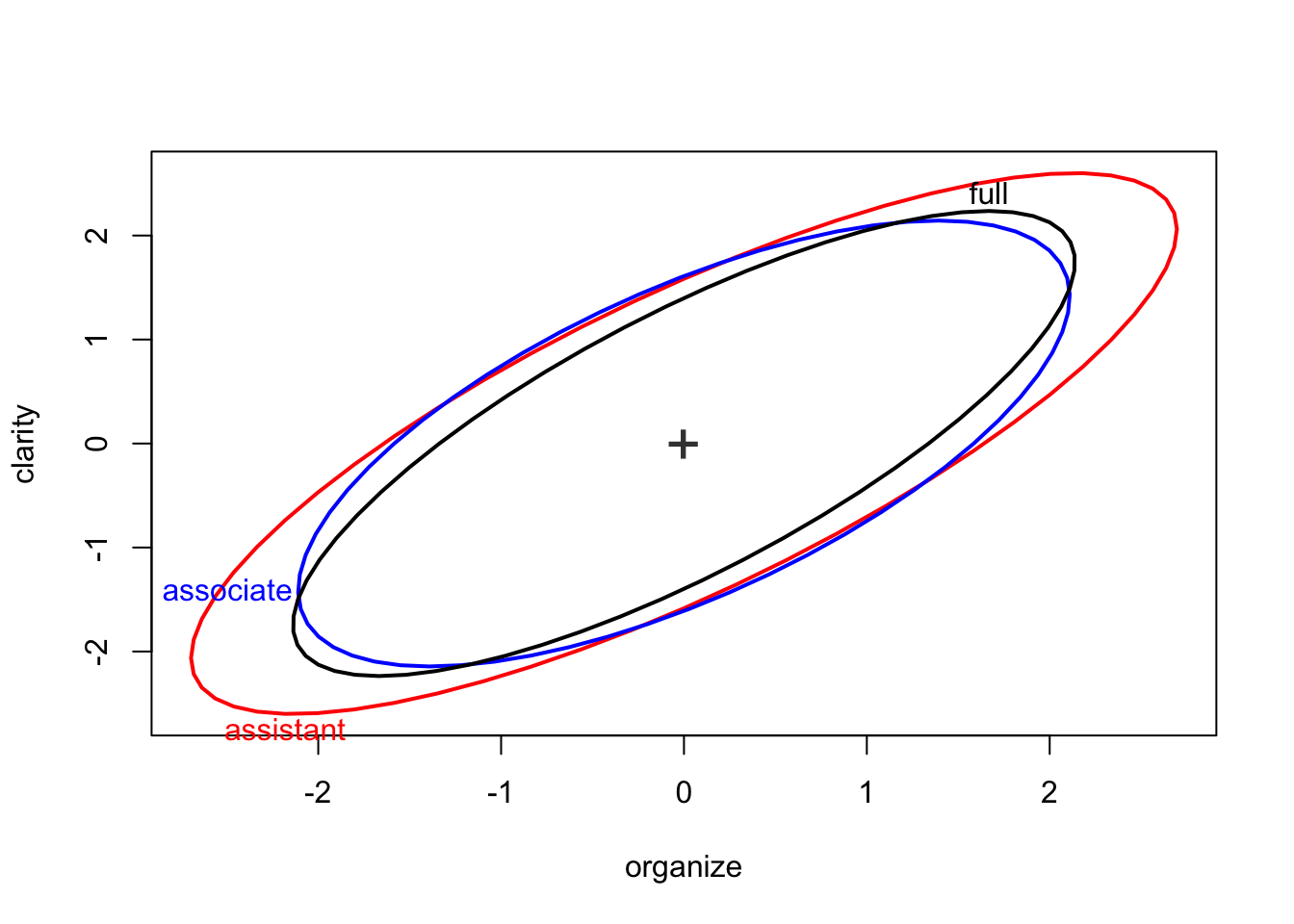
We can even add the pooled covariance ellipse, centered or uncentered.
covEllipses(Teacher_ratings[,1:6], Teacher_ratings$group,
fill=c(rep(FALSE,3), TRUE))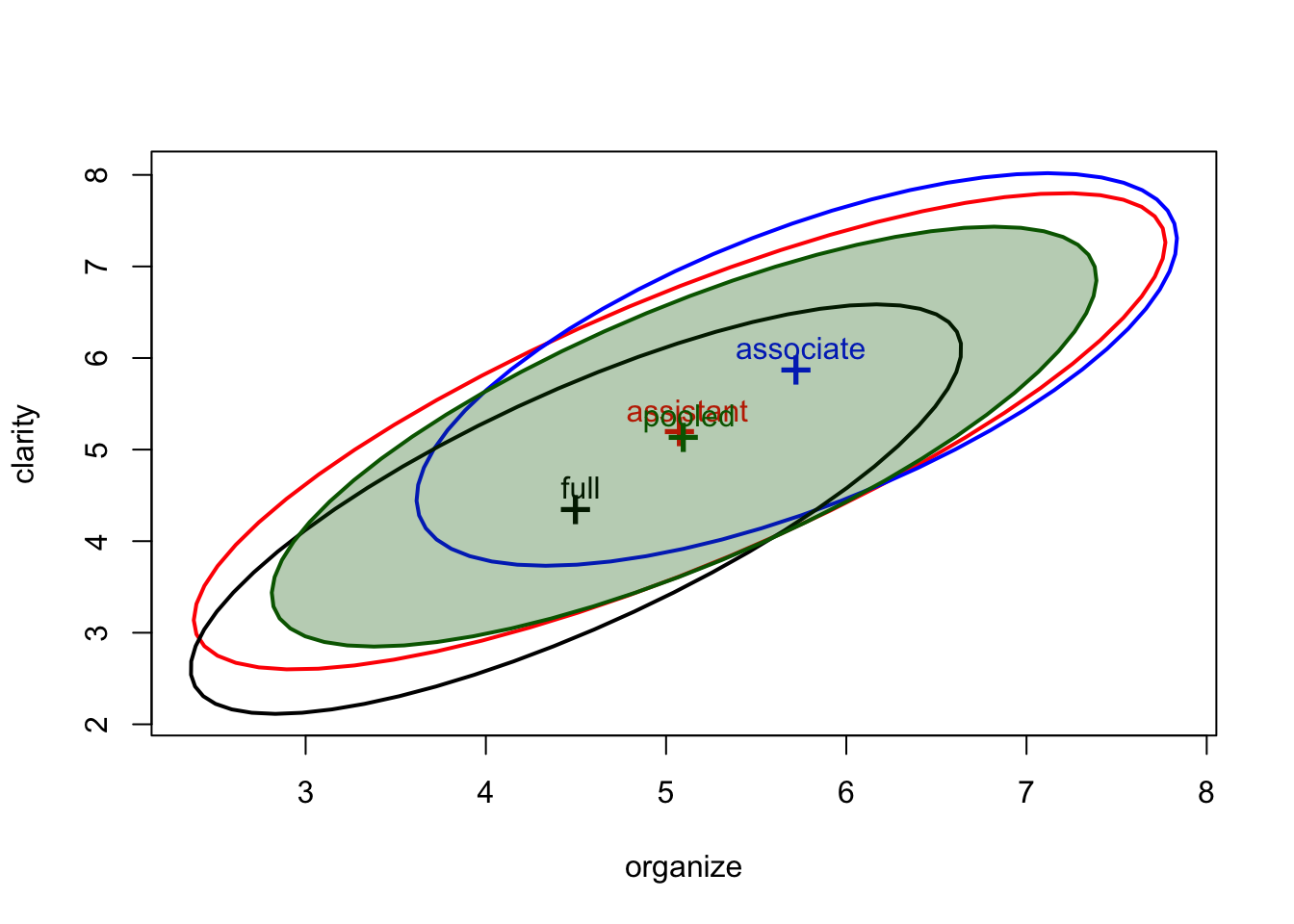
covEllipses(Teacher_ratings[,1:6], Teacher_ratings$group,center=TRUE, fill=c(rep(FALSE,3), TRUE), fill.alpha=.1, label.pos=c(1:3,0))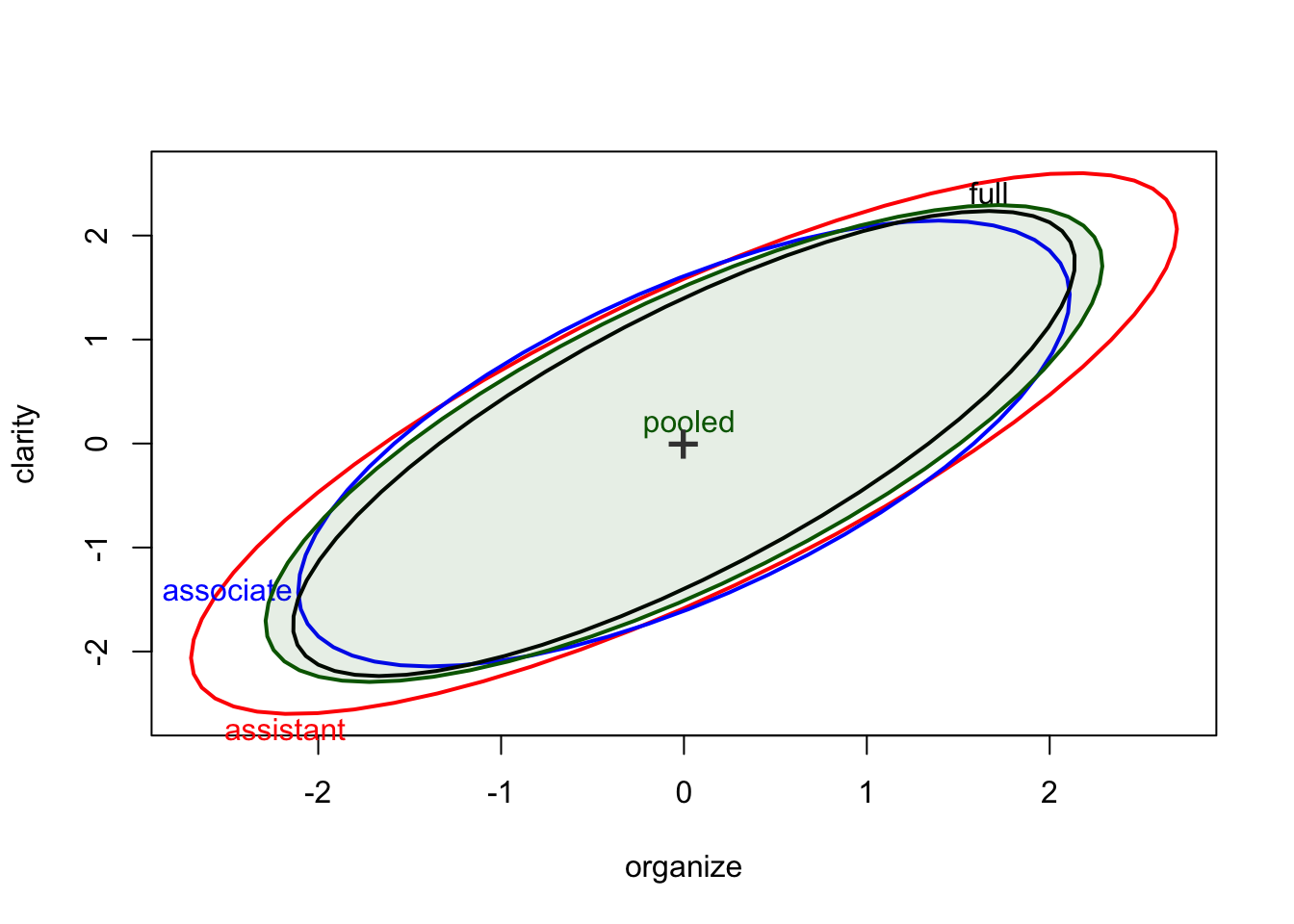
Let’s see the covariances between all possible pairs of the variables.
covEllipses(Teacher_ratings[,1:6], Teacher_ratings$group,
fill=c(rep(FALSE,3), TRUE), variables=1:6,
fill.alpha=.1,pooled=F)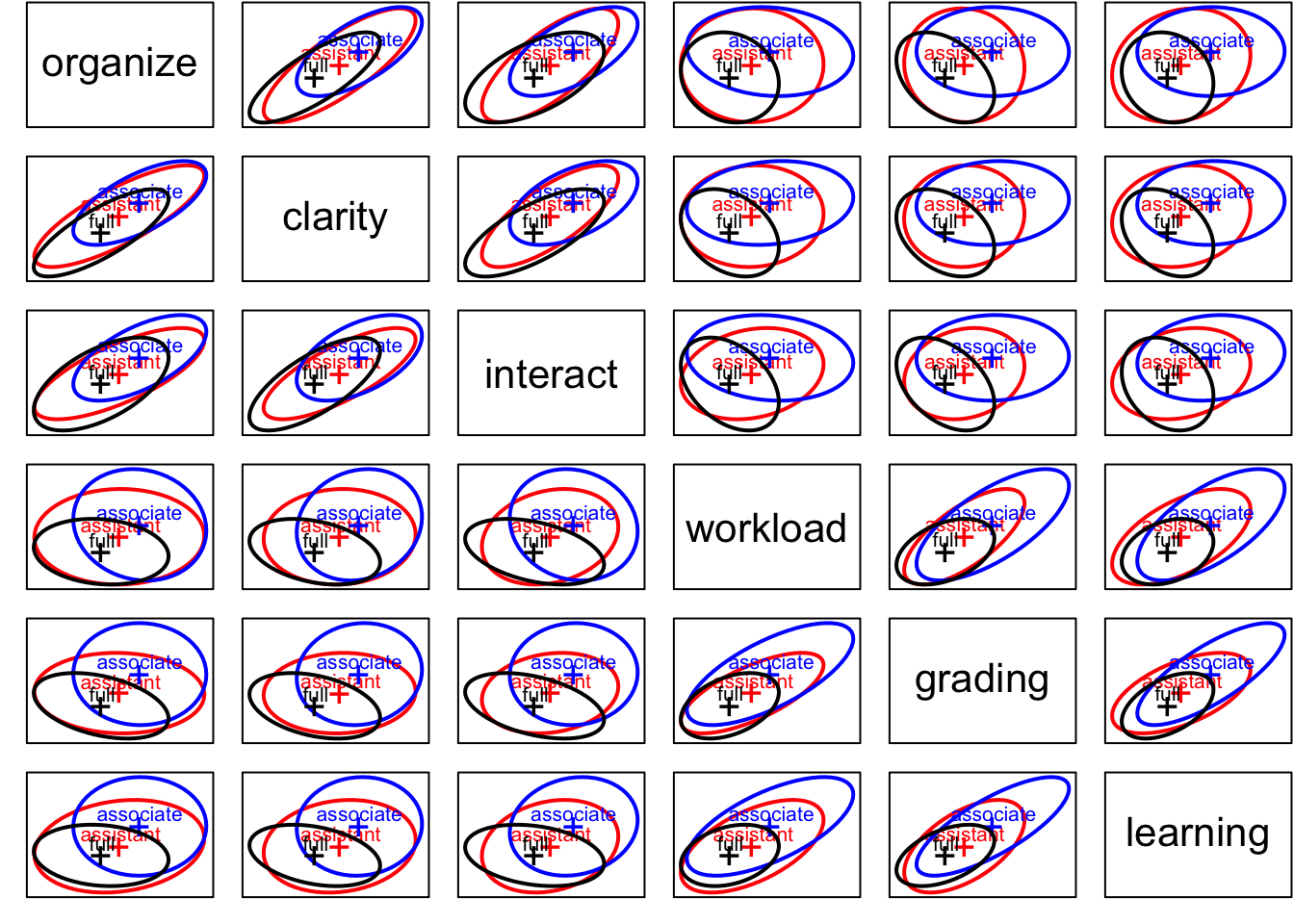
covEllipses(Teacher_ratings[,1:6], Teacher_ratings$group, center=T, fill=c(rep(FALSE,3), TRUE), variables=1:6,
fill.alpha=.1,pooled=F)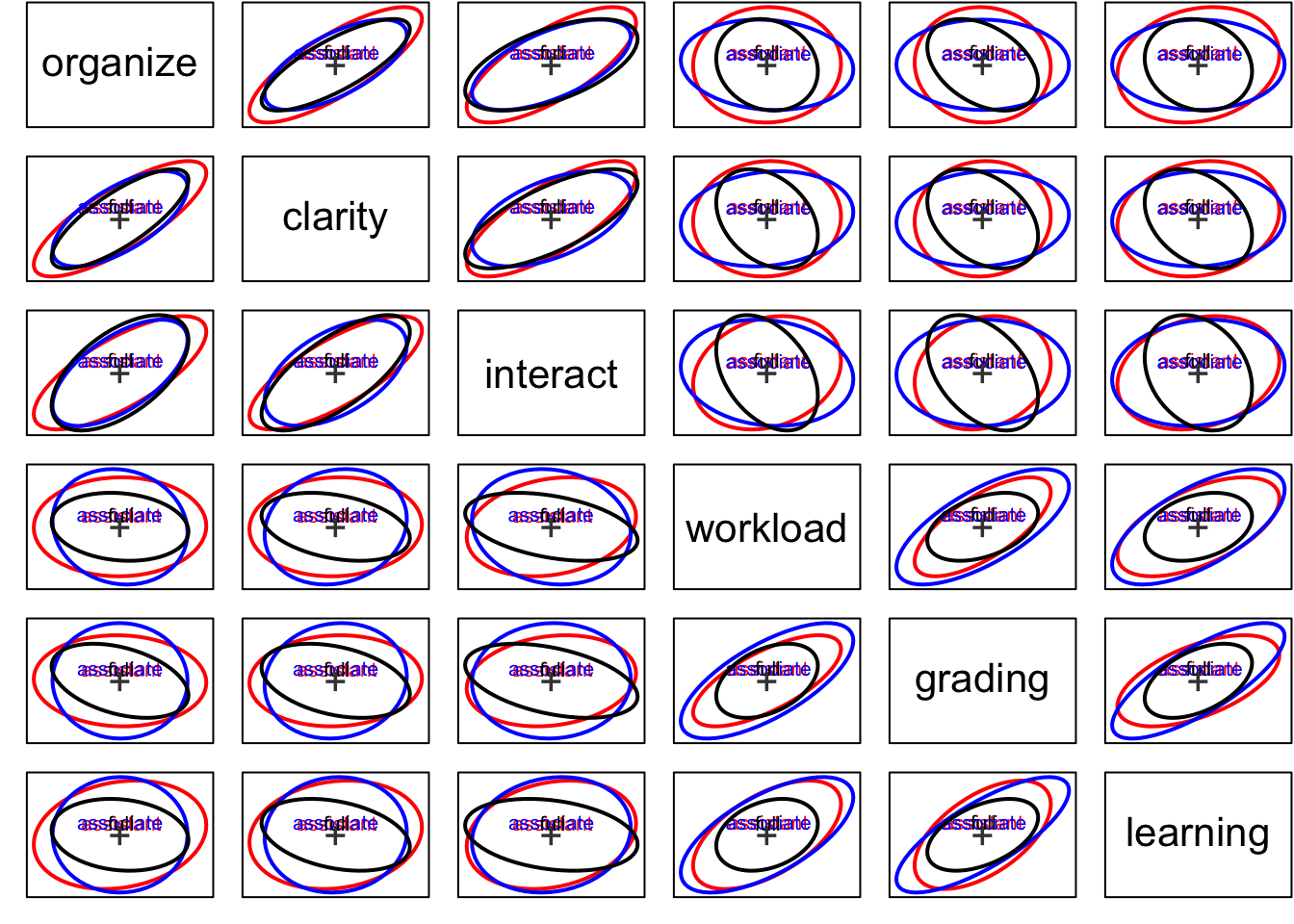
We can also view the data points in principal components space.
manova.data3.pca <- prcomp(Teacher_ratings[,1:6])
summary(manova.data3.pca)## Importance of components:
## PC1 PC2 PC3 PC4 PC5 PC6
## Standard deviation 2.6597 2.2185 1.02483 0.92508 0.80284 0.68122
## Proportion of Variance 0.4713 0.3279 0.06997 0.05701 0.04294 0.03092
## Cumulative Proportion 0.4713 0.7992 0.86913 0.92614 0.96908 1.00000op <- par(mfcol=c(1,2), mar=c(5,4,1,1)+.1)
covEllipses(manova.data3.pca$x, Teacher_ratings$group,
fill=c(rep(FALSE,3), TRUE),
label.pos=1:4, fill.alpha=.1, asp=1,pooled=F)
covEllipses(manova.data3.pca$x, Teacher_ratings$group,
fill=c(rep(FALSE,3), TRUE), variables=3:4,
label.pos=1:4, fill.alpha=.1, asp=1,pooled=F)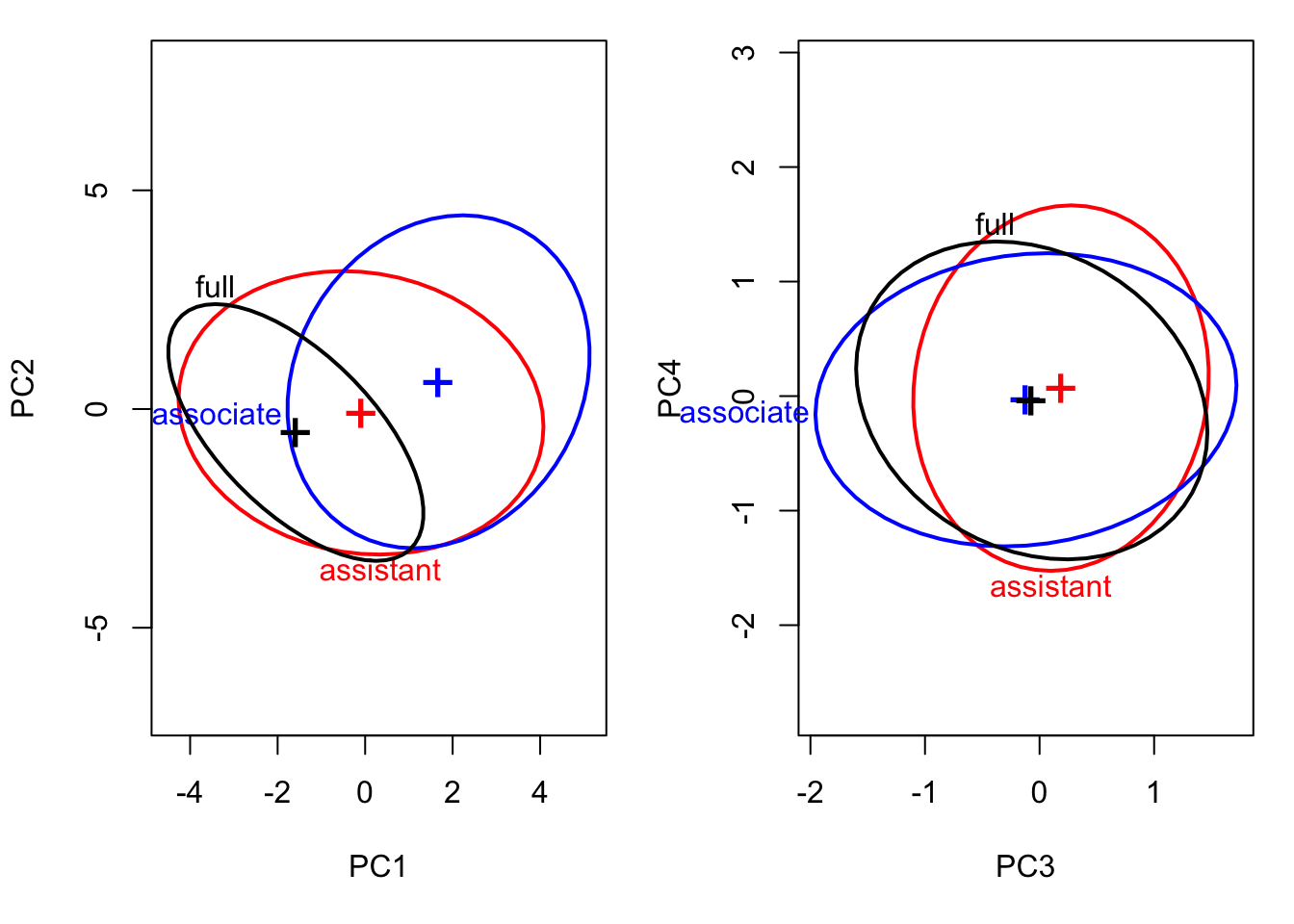
covEllipses(manova.data3.pca$x, Teacher_ratings$group,
fill=c(rep(FALSE,3), TRUE), variables=1:6,
label.pos=1:4, fill.alpha=.1, asp=1,pooled=F)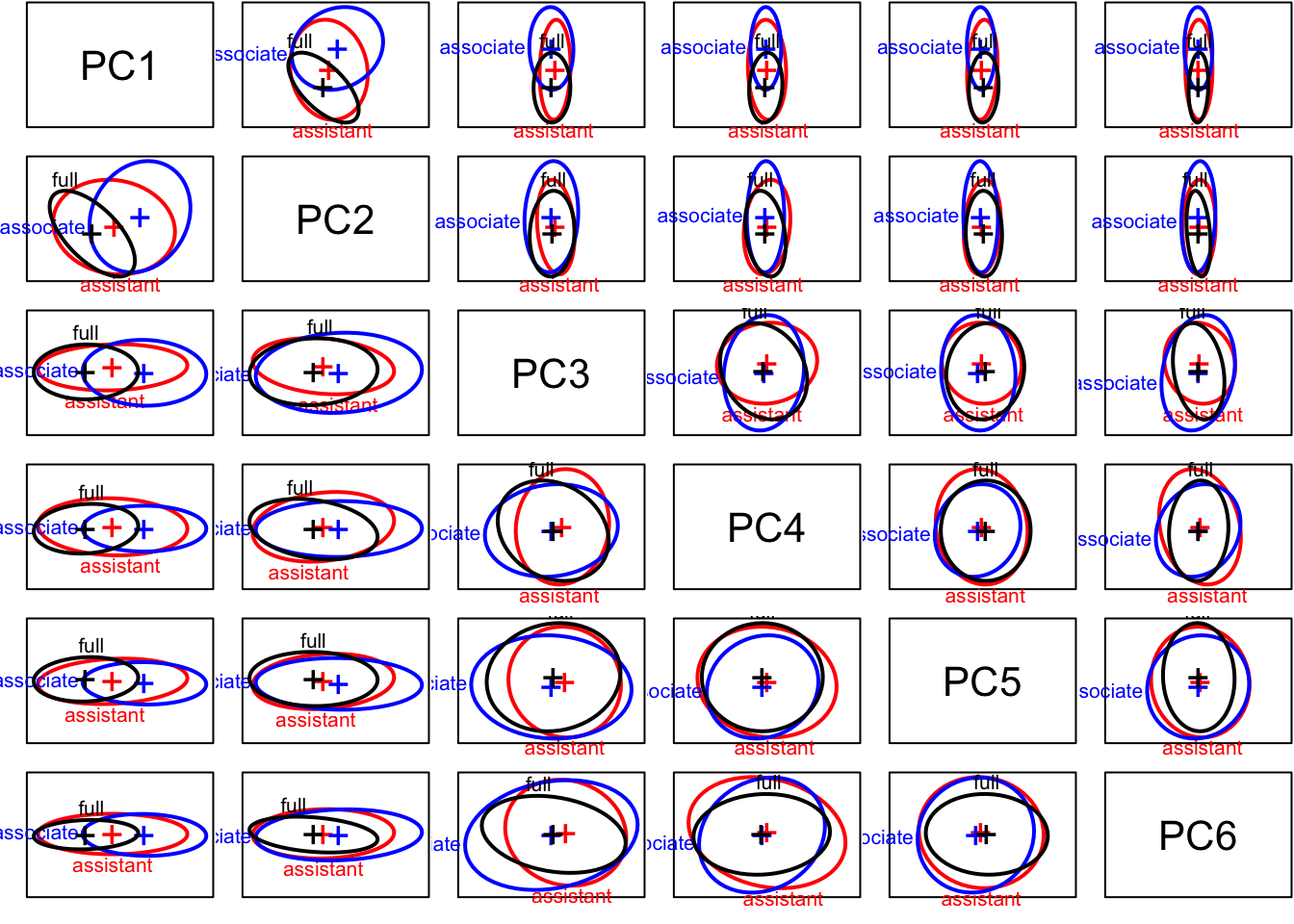
par(op)Step 3: Implement MANOVA
Multivariate tests
manova.model3 <- manova(cbind(organize, clarity, interact, workload, grading, learning) ~ group, Teacher_ratings)
summary(manova.model3, test = "Pillai", intercept=T)## Df Pillai approx F num Df den Df Pr(>F)
## (Intercept) 1 0.96611 532.21 6 112 < 2.2e-16 ***
## group 2 0.33405 3.78 12 226 3.086e-05 ***
## Residuals 117
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(manova.model3, test = "Wilks", intercept=T)## Df Wilks approx F num Df den Df Pr(>F)
## (Intercept) 1 0.03389 532.21 6 112 < 2.2e-16 ***
## group 2 0.67235 4.10 12 224 8.686e-06 ***
## Residuals 117
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(manova.model3, test = "Hotelling-Lawley", intercept=T)## Df Hotelling-Lawley approx F num Df den Df Pr(>F)
## (Intercept) 1 28.5113 532.21 6 112 < 2.2e-16 ***
## group 2 0.4778 4.42 12 222 2.452e-06 ***
## Residuals 117
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(manova.model3, test = "Roy", intercept=T)## Df Roy approx F num Df den Df Pr(>F)
## (Intercept) 1 28.511 532.21 6 112 < 2.2e-16 ***
## group 2 0.457 8.61 6 113 1.038e-07 ***
## Residuals 117
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Box’s M test
library(heplots)
boxtest6 <- boxM(Teacher_ratings[,1:6], Teacher_ratings$group)
summary(boxtest6)## Summary for Box's M-test of Equality of Covariance Matrices
##
## Chi-Sq: 56.1123
## df: 42
## p-value: 0.07135
##
## log of Covariance determinants:
## assistant associate full pooled
## 2.05352330 2.25782486 -0.03899053 1.94116978
##
## Eigenvalues:
## assistant associate full pooled
## 1 7.5445874 6.6965430 5.9976581 5.7899999
## 2 4.4509180 4.5908871 1.4803104 4.3594037
## 3 1.1212748 1.4661925 1.0181861 1.0496581
## 4 0.6645808 0.7631897 0.6961131 0.8676318
## 5 0.6485849 0.5527171 0.6224494 0.6427895
## 6 0.4803105 0.5029064 0.2455394 0.4714999
##
## Statistics based on eigenvalues:
## assistant associate full pooled
## product 7.7953180 9.5622673 0.9617598 6.9668960
## sum 14.9102564 14.5724359 10.0602564 13.1809829
## precision 0.1567995 0.1624123 0.1118546 0.1617085
## max 7.5445874 6.6965430 5.9976581 5.7899999Step 4: Follow-up analyses
Univariate ANOVA
anova(lm(organize~group, Teacher_ratings))## Analysis of Variance Table
##
## Response: organize
## Df Sum Sq Mean Sq F value Pr(>F)
## group 2 30.05 15.0250 6.4928 0.002118 **
## Residuals 117 270.75 2.3141
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1anova(lm(clarity~group, Teacher_ratings))## Analysis of Variance Table
##
## Response: clarity
## Df Sum Sq Mean Sq F value Pr(>F)
## group 2 46.717 23.3583 10.052 9.362e-05 ***
## Residuals 117 271.875 2.3237
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1anova(lm(interact~group, Teacher_ratings))## Analysis of Variance Table
##
## Response: interact
## Df Sum Sq Mean Sq F value Pr(>F)
## group 2 42.65 21.3250 8.0413 0.0005343 ***
## Residuals 117 310.27 2.6519
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1anova(lm(workload~group, Teacher_ratings))## Analysis of Variance Table
##
## Response: workload
## Df Sum Sq Mean Sq F value Pr(>F)
## group 2 36.6 18.3000 7.9095 6e-04 ***
## Residuals 117 270.7 2.3137
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1anova(lm(grading~group, Teacher_ratings))## Analysis of Variance Table
##
## Response: grading
## Df Sum Sq Mean Sq F value Pr(>F)
## group 2 45.60 22.8000 13.519 5.224e-06 ***
## Residuals 117 197.32 1.6865
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1anova(lm(learning~group, Teacher_ratings))## Analysis of Variance Table
##
## Response: learning
## Df Sum Sq Mean Sq F value Pr(>F)
## group 2 42.45 21.225 11.224 3.473e-05 ***
## Residuals 117 221.25 1.891
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Tukey’s HSD test
library(agricolae)
(HSD.test(aov(organize~group, Teacher_ratings), "group"))$groups## organize groups
## associate 5.725 a
## assistant 5.075 ab
## full 4.500 b(HSD.test(aov(clarity~group, Teacher_ratings), "group"))$groups ## clarity groups
## associate 5.875 a
## assistant 5.200 a
## full 4.350 b(HSD.test(aov(interact~group, Teacher_ratings), "group"))$groups ## interact groups
## associate 5.750 a
## assistant 4.875 b
## full 4.300 b(HSD.test(aov(workload~group, Teacher_ratings), "group"))$groups ## workload groups
## associate 5.00 a
## assistant 4.40 ab
## full 3.65 b(HSD.test(aov(grading~group, Teacher_ratings), "group"))$groups ## grading groups
## associate 5.025 a
## assistant 4.125 b
## full 3.525 b(HSD.test(aov(learning~group, Teacher_ratings), "group"))$groups ## learning groups
## associate 5.250 a
## assistant 4.275 b
## full 3.825 b“Manual” multivariate multiple comparisons
summary(manova(cbind(organize, clarity, interact, workload, grading, learning) ~ group, Teacher_ratings[Teacher_ratings$group %in% c("assistant","associate"), ]), intercept = T)## Df Pillai approx F num Df den Df Pr(>F)
## (Intercept) 1 0.96255 312.683 6 73 < 2e-16 ***
## group 1 0.15791 2.282 6 73 0.04494 *
## Residuals 78
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(manova(cbind(organize, clarity, interact, workload, grading, learning) ~ group, Teacher_ratings[Teacher_ratings$group %in% c("assistant","full"),]), intercept = T)## Df Pillai approx F num Df den Df Pr(>F)
## (Intercept) 1 0.96638 349.77 6 73 < 2e-16 ***
## group 1 0.16240 2.36 6 73 0.03876 *
## Residuals 78
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(manova(cbind(organize, clarity, interact, workload, grading, learning) ~ group, Teacher_ratings[Teacher_ratings$group %in% c("full","associate"),]), intercept = T)## Df Pillai approx F num Df den Df Pr(>F)
## (Intercept) 1 0.97224 426.17 6 73 < 2.2e-16 ***
## group 1 0.44391 9.71 6 73 7.584e-08 ***
## Residuals 78
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Discriminant function analysis (Canonical discrimination analysis/linear discriminant analysis)
library(candisc)
disc <- candisc(manova(cbind(organize, clarity, interact, workload, grading, learning) ~ group, Teacher_ratings))
summary(disc, scores = F)##
## Canonical Discriminant Analysis for group:
##
## CanRsq Eigenvalue Difference Percent Cumulative
## 1 0.313654 0.45699 0.43617 95.6427 95.643
## 2 0.020395 0.02082 0.43617 4.3573 100.000
##
## Class means:
##
## Can1 Can2
## assistant -0.068936 0.200953
## associate 0.849812 -0.087734
## full -0.780876 -0.113219
##
## std coefficients:
## Can1 Can2
## organize 0.094533 -0.19892
## clarity 0.400276 0.85437
## interact 0.238116 -0.47068
## workload 0.084000 0.86171
## grading 0.550587 -0.25997
## learning 0.223505 -0.68948- Wilks’ Lambda
Wilks(disc)##
## Test of H0: The canonical correlations in the
## current row and all that follow are zero
##
## LR test stat approx F numDF denDF Pr(> F)
## 1 0.67235 4.0984 12 224 8.686e-06 ***
## 2 0.97960 0.4705 5 113 0.7975
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1- Structure coefficients
disc$structure## Can1 Can2
## organize 0.5639597 0.0835453
## clarity 0.6771253 0.3721678
## interact 0.6200649 -0.1114412
## workload 0.6104303 0.3303997
## grading 0.7729296 -0.1269871
## learning 0.7097412 -0.3822816coef(disc, type = "structure")## Can1 Can2
## organize 0.5639597 0.0835453
## clarity 0.6771253 0.3721678
## interact 0.6200649 -0.1114412
## workload 0.6104303 0.3303997
## grading 0.7729296 -0.1269871
## learning 0.7097412 -0.3822816- Plot the discriminant function space
plot(disc, which = 1:2, conf = 0.95,asp = 1, scale=4,
var.col = "blue", var.lwd = par("lwd"), var.cex = 1,
rev.axes=c(FALSE, FALSE),
ellipse=FALSE, ellipse.prob = 0.68, fill.alpha=0.1,
prefix = "DF", suffix=TRUE,
titles.1d = c("Canonical scores", "Structure"))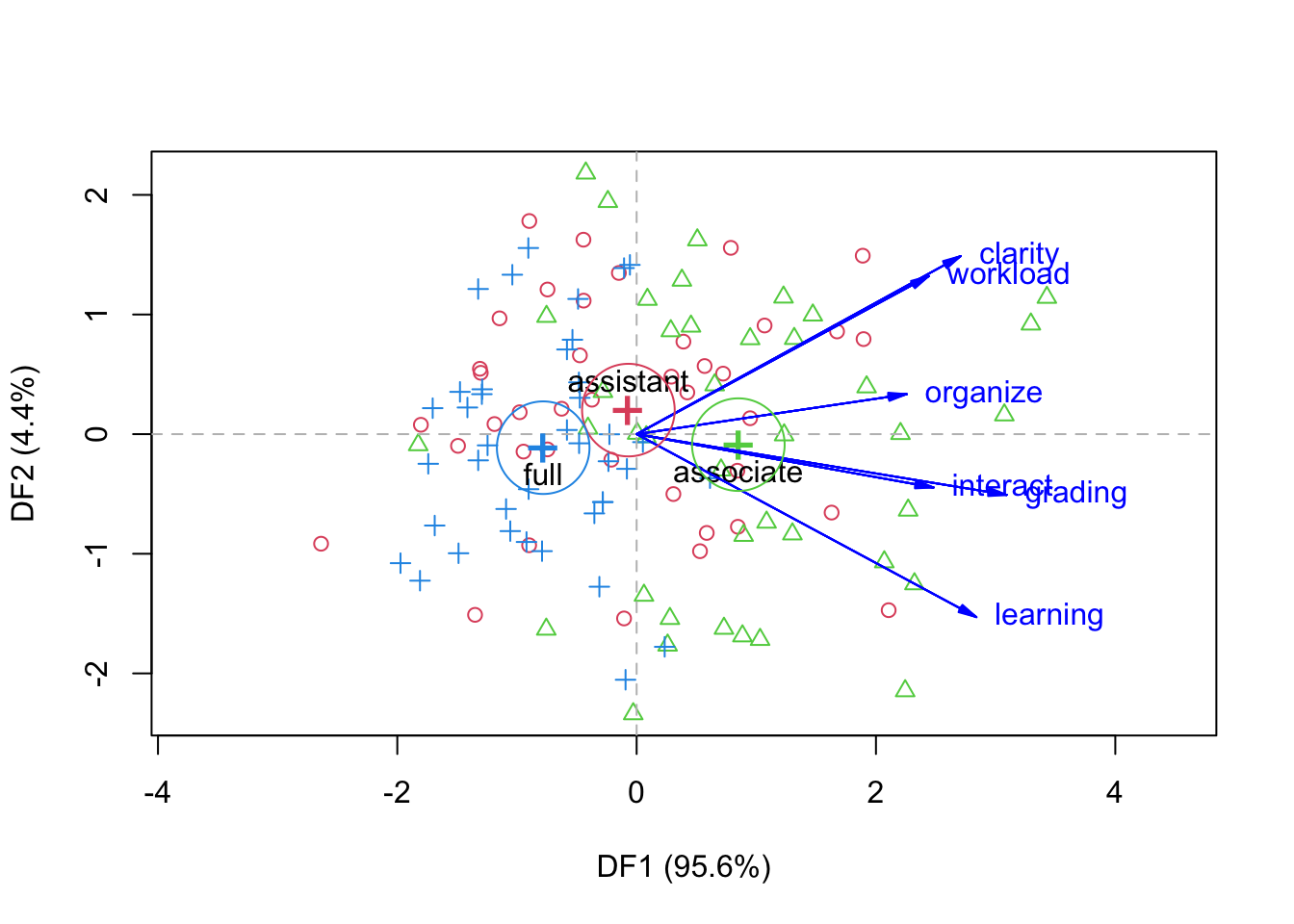
- Examine the distribution of LDA scores
Teacher_ratings$lda1 <- disc$scores[,2]
Teacher_ratings$lda2 <- disc$scores[,3]
ggplot(Teacher_ratings, aes(lda1, fill =group)) + geom_density(alpha = 0.5, color = NA) + theme(legend.position = "top")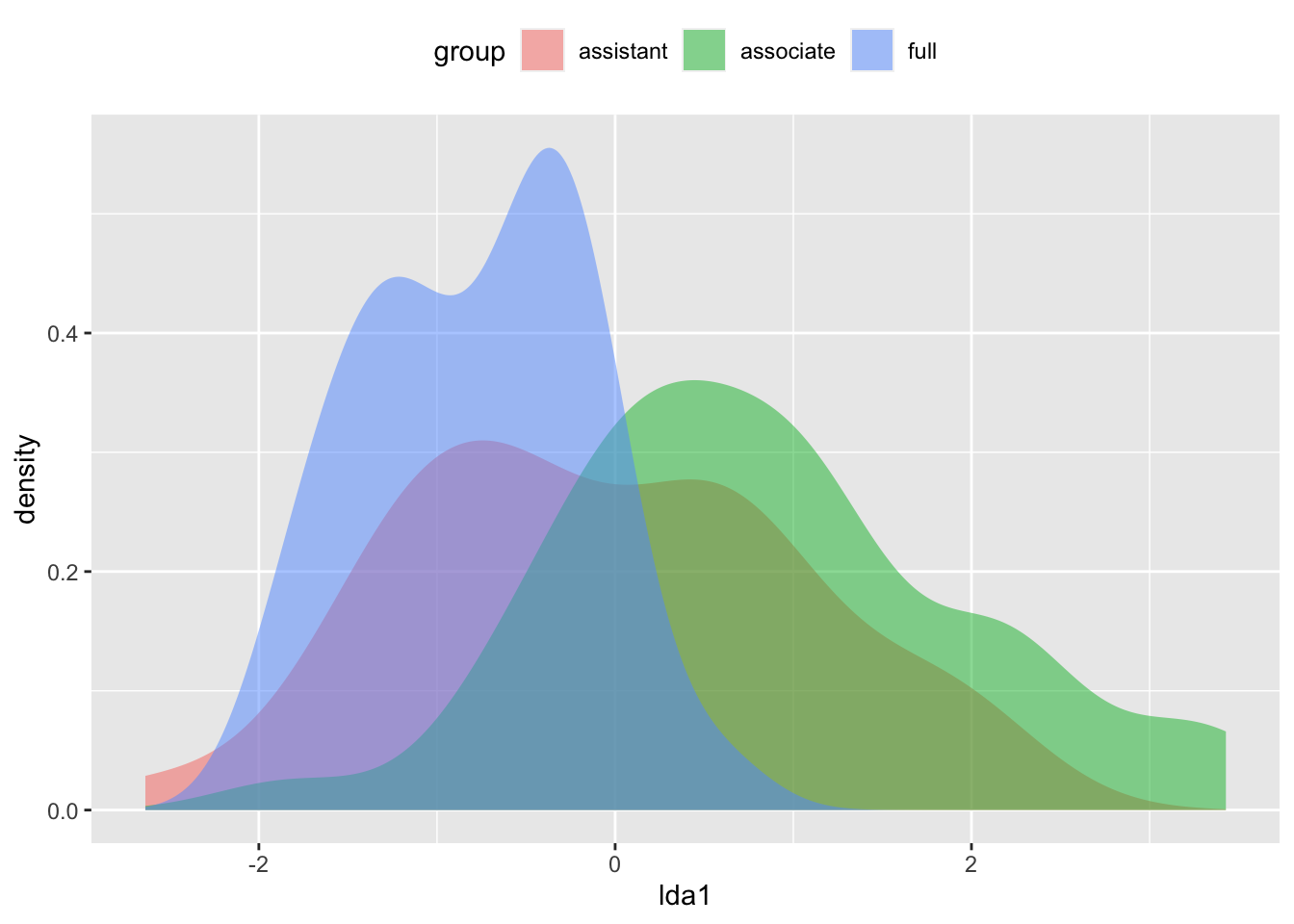
ggplot(Teacher_ratings, aes(lda2, fill =group)) + geom_density(alpha = 0.5, color = NA) + theme(legend.position = "top")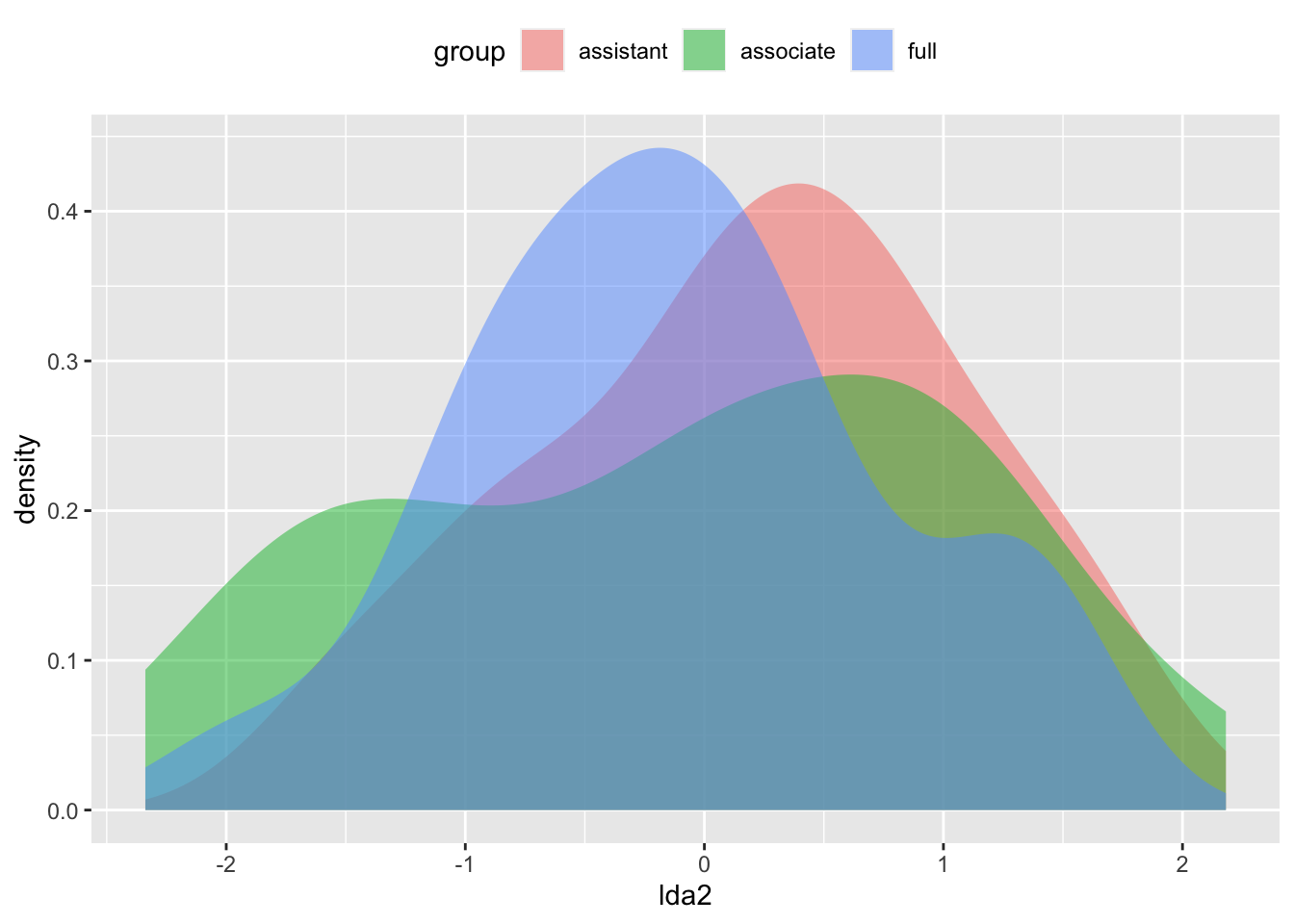
rc_plot3 <- ggplot(data = Teacher_ratings, aes(x=group, y=lda1, fill = group)) +
geom_flat_violin(position = position_nudge(x = .2, y = 0), alpha = .8) +
geom_point(aes(y=lda1, color = group), position = position_jitter(width = .15), size = .5, alpha = 0.8) +
geom_boxplot(width = .1, guides = FALSE, outlier.shape = NA, alpha = 0.5) +
expand_limits(x = 4, y = 4) +
ylab("First Discriminant Function") +
guides(fill = FALSE) +
guides(color = FALSE) +
scale_color_brewer(palette = "Set1") +
scale_fill_brewer(palette = "Set1") +
theme_bw() +
raincloud_theme
rc_plot3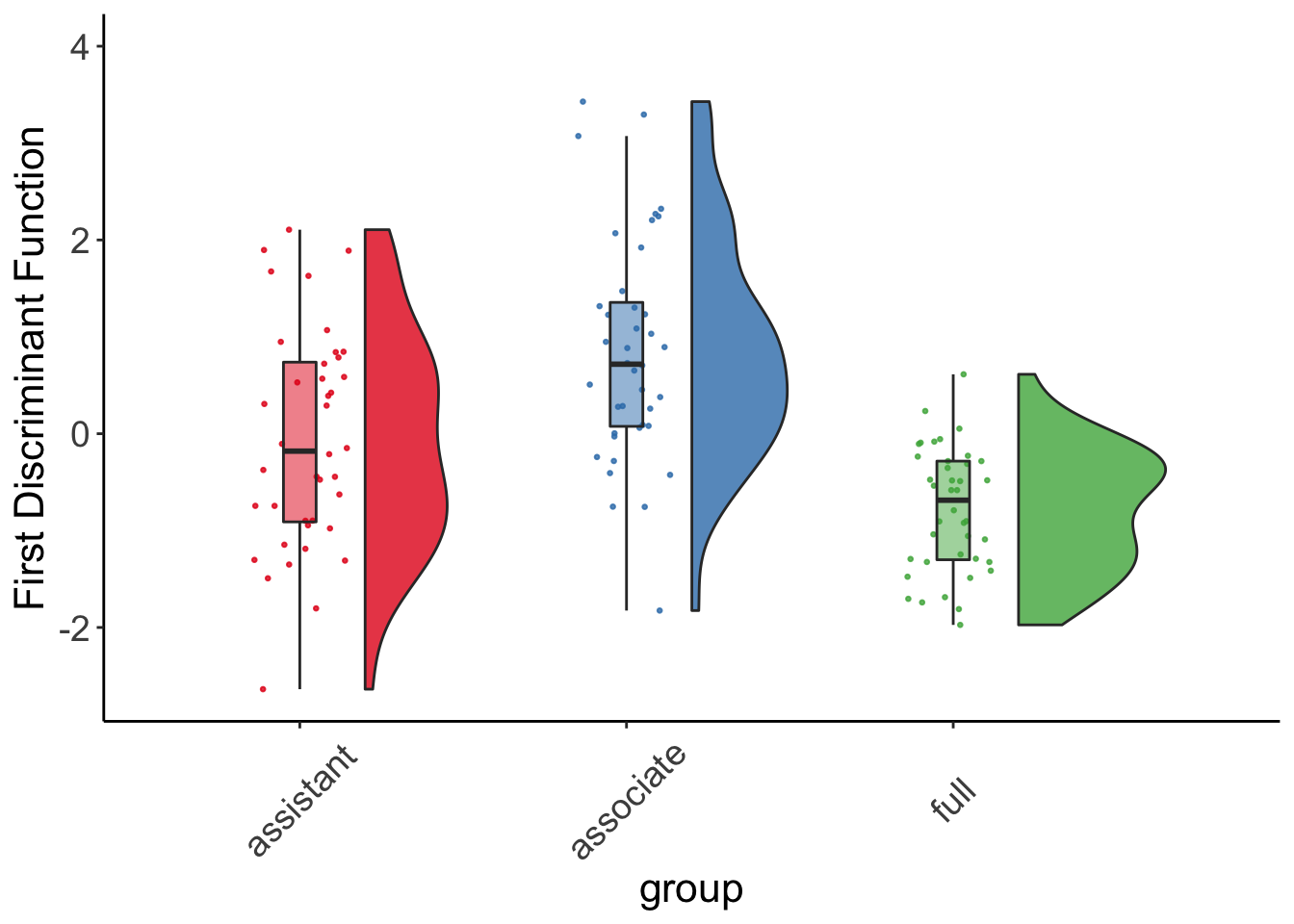
The R script for this tutorial can be found here.
© Copyright 2022 Yi Feng and Gregory R. Hancock.